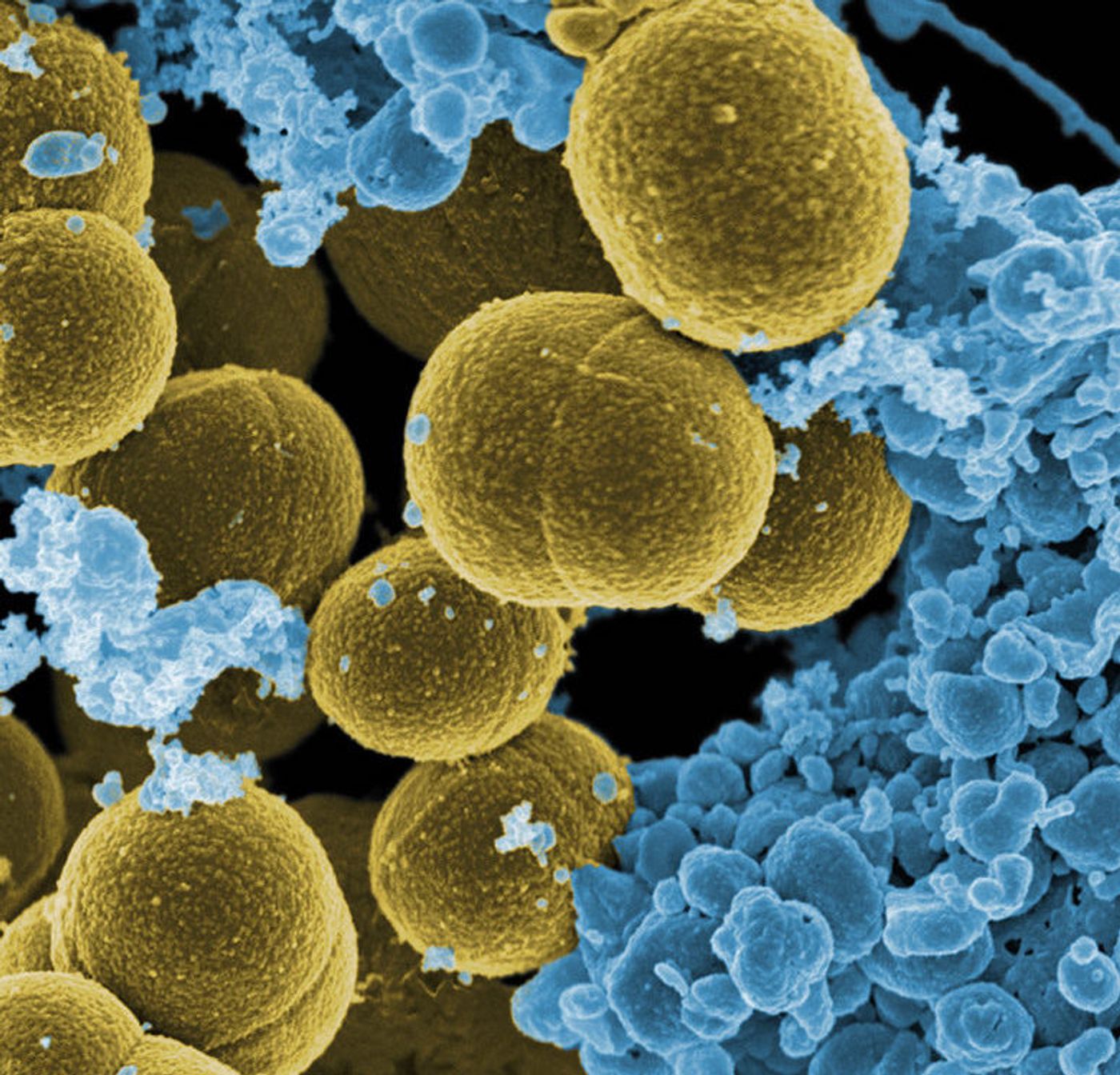
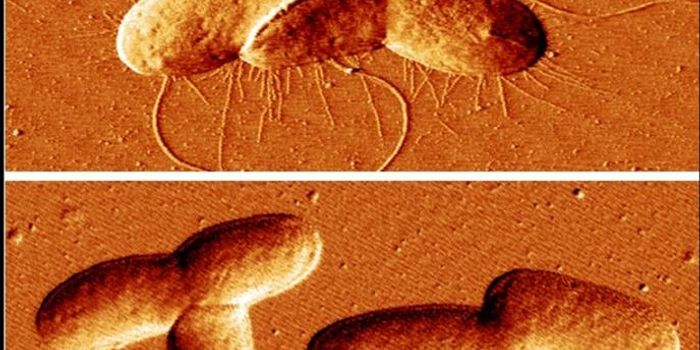
Current hospital laboratory testing is not great at identifying bacterial infections; it can take precious time and resources to learn what microbe is causing the illness, and what treatment to use. A new technology created at Rice University aims to solve that problem. They have created a "universal microbial diagnostic" (UMD), which is able to quickly pinpoint hundreds of pathogenic bacteria easily and at low cost. The technology utilizes bits of randomly assembled DNA in combination with mathematical tools that were first created for digital phones and cameras.
The
research paper about the UMD was published in Science Advances by the Rice team. They show that UMD can verifiable identify 11 strains of known bacteria with the same set of five DNA probes. Those probes are not disease-specific. Instead, the tool uses genetics to indicate bacterial identity without needing a unique DNA test to probe for individual pathogens.
"If a laboratory today wants to test for 200 known pathogenic species, they need 200 different tests, each with its own specific DNA probe that was designed specifically to bind with DNA from a particular pathogen," explained Richard Baraniuk, the lead scientist on the new study. "Our technology is fundamentally different. With a small set of DNA probes, we can test for a large number of species."
"Probe A is only good for finding bacterium A, and probe B is only good for finding bacterium B," said the lead author of the work, Amirali Aghazadeh, a graduate student in electrical and computer engineering in Baraniuk's lab. "Manufacturing such a probe for a new disease can take days to months and also requires expensive facilities that are only available in developed countries. With universal microbial diagnostic, we won't need new probes or to change any other parts of the sensing hardware," he continued. "For any newly discovered bacterial strain, we can just adapt the software a little bit, and then the same platform can identify the new bacterium like any other."
Their report features several computer simulations, with one illustrating how five probes can identify 40 different bacterial strains. Another shows that the technology is able to tell the difference between 24 various strains of Staphylococcus bacteria.
The spread of antibiotic-resistance bacteria is a rising challenge for public health and is critical to limit in hospitals. The researchers hope this new tool will help solve that problem.
"In many U.S. hospitals, it still takes several days to definitively identify the specific bacterium that's making someone sick," commented Baraniuk, the Victor E. Cameron Professor of Electrical and Computer Engineering at Rice. "The lack of rapid bacterial diagnostics can promote antibiotic resistance. Having an accurate, efficient and rapid system for identifying infectious pathogens quickly and inexpensively would help, and such a system would also be a valuable tool for public health, defense, global health and environmental science."
In times gone by, bacteria had to cultured to be identified which was a laborious task. Genomic technologies made testing much easier, but it still requires expensive equipment and specific probes to identify bacterial strains.
UMD doesn’t rely on probes that require exact specificity to find their target. Instead, the system learns how well random pieces of DNA bind to the target DNA. The math that is then applied is called compressive sensing. Thus, the binding characteristics of probe to target are assessed to create a profile for the organism under examination. Deductive reasoning is then used to find the matching pathogen.
"We believe the system will be most useful for rapidly and accurately looking for known targets, such as any organism on the World Health Organization's list of known pathogens, but another advantage of this method is that we get useful information even for organisms that have never been sequenced before," commented a study co-author Rebekah Drezek, Professor of Bioengineering and of Electrical and Computer Engineering. "We'd be able to tell what pathogen a new disease is most closely related to."
If you'd like to know more about compressive sensing, a lecture given by Professor Baraniuk is shown in the video above from the University of Delaware.
Sources:
Phys.org via
Rice University,
Science Advances