Scientists would like to get a better look at soil microbes in order to have a better understanding of how they impact our planet and ecosystem. There is a dearth of knowledge about the molecular characteristics of their roles in and responses to environmental dynamics. Researchers have now
reported in the journal mSystems one of the most detailed analysis seen yet of a soil metagenome, the genetic composition of a soil sample, as well as the first ever complete genome of a microbe taken from a complex soil sample.
Microbes in soil are an important part of our ecosystem; they are participants in the cycling of carbon as well as other nutrients. They also aid in plant growth and influence how plants absorb nutrients. Soil microbes can have a big effect on how the planet stores and releases carbon.
"We're trying to sort out the broad questions. What are the various microbes in the microbial community doing? Which species are very active and which seem dormant? How do they all fit together?" explained Janet Jansson, a microbiologist and the senior author.
Soil microorganisms present researchers with many challenges. Soil samples mean a messy mix of material that has been subject to many detrimental forces. Soil microbes are difficult to grow in the lab, so while investigators have learned a lot about the species that are present in soil samples, they have not been able to get a good look at how they interact.
"Imagine taking a thick book written in hundreds of different languages, chopping the book up into pieces the size of grains of rice, and then having to put it back together again," said Richard Allen White III, a post-doctoral associate and first author of the paper. "That's not unlike the challenge we face when we try to understand what's going on in even a handful of soil."
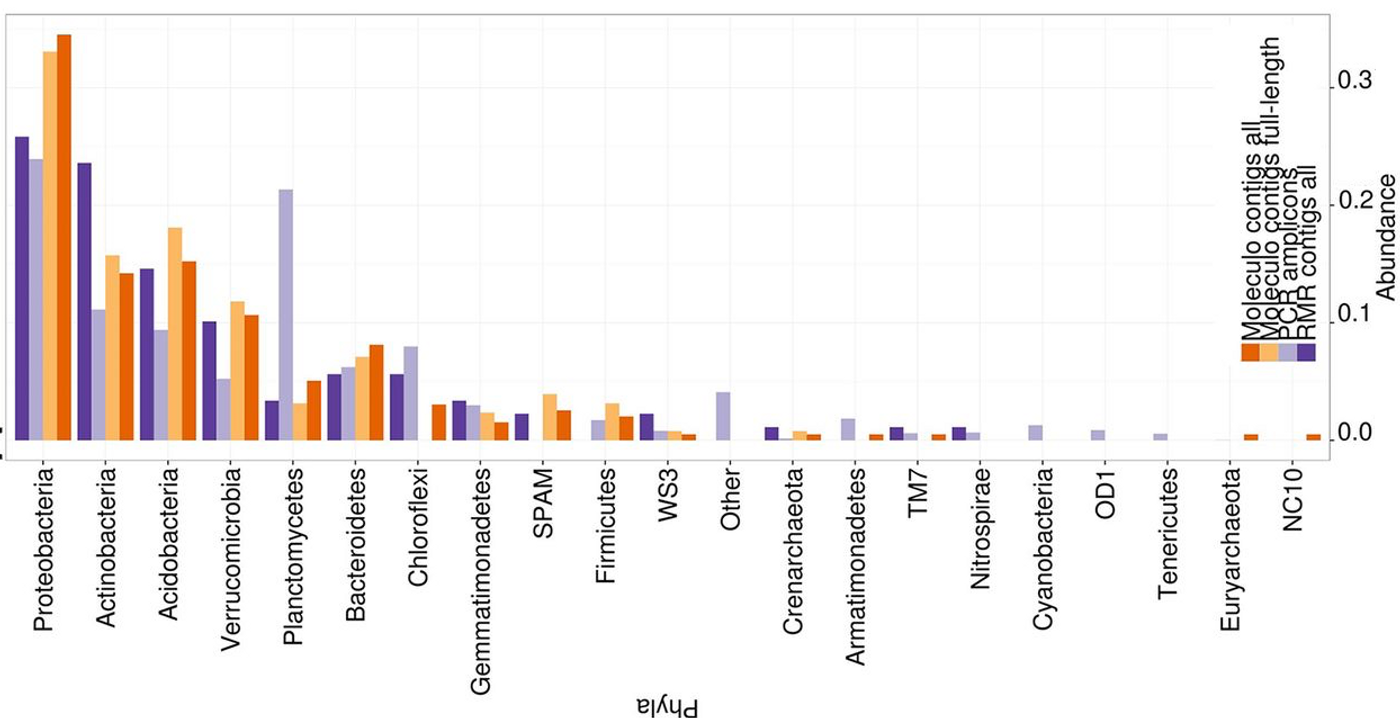
There are as many as 50 to 100 times the microbial species in a typical soil sample compared to a sample from the human gut. The researchers applied Moleculo synthetic long-read sequencing technology to their study in combination with other methods, and using powerful supercomputers at the Environmental Molecular Sciences Laboratory on the campus of the Pacific Northwest National Laboratory they were able to achieve their goal.
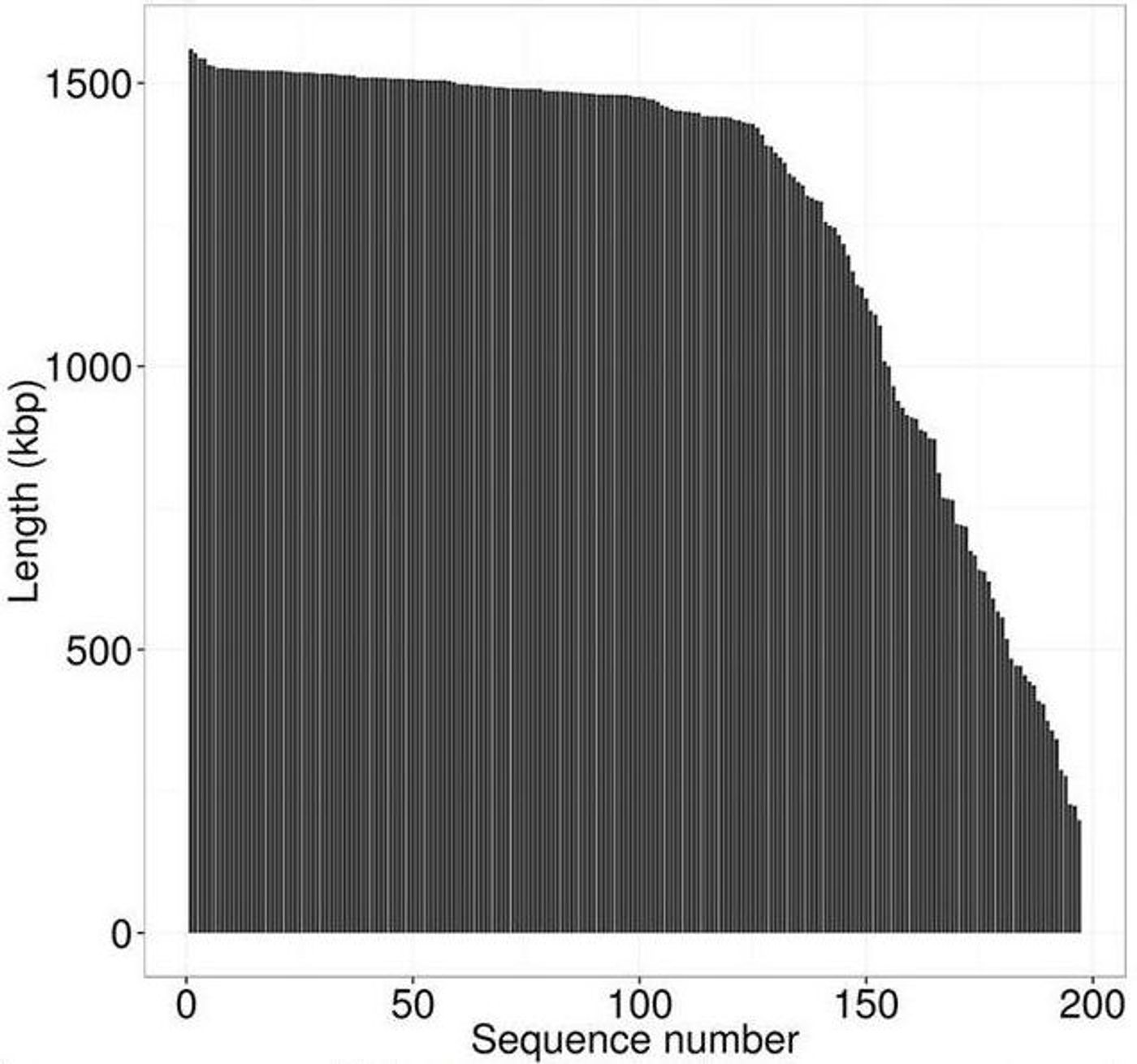
To perform the study, White took adavantage of a sequencing technology that had been developed in the laboratory of his former adviser, Stephen Quake of Stanford University. The research team broke down the DNA into small fragments and used the computational tools to build it up. They got 10,000 pieces of DNA, and with each being longer than 10 kilobases. Other attempts at soil metagenome sequencing have yielded numbers such as nine pieces of DNA of that length.
"Soil is one of the most complex and diverse ecosystems on the planet. It's a complex three-dimensional substrate; there's nothing else quite like it," commented White.
"We're at the point where we've put together a few long sentences of a very large book," he continued. "We've gone from having a few words or parts of words to having a few sentences. But we've got a long ways to go. We are in our baby steps of identifying who's in there and what they're doing."
Want to know more about the microbes that live in soil and how they benefit people? Check out the video below from the USDA's Agricultural Research Service.
A more technical discussion of soil microbes is shown in the video below.
Sources:
Science Daily via
Pacific Northwest National Laboratory,
mSystems