Scientists have begun to voice concerns about a perceived lack of data reproducibility. A pillar of the scientific method, data should be reproducible to verify its authenticity and build additional research on a solid foundation.
Two new research papers have been published in the Open Access journal
GigaScience, in collaboration with
protocols.io, which describes itself as “An up-to-date open access repository of science methods and a collaborative protocol-centered platform.” The research papers concern two parasitic organisms, tapeworm and scabies, and the collaborative nature of the publication highlights a new system for sharing methodology that could help scientists repeat and expand on complex work.
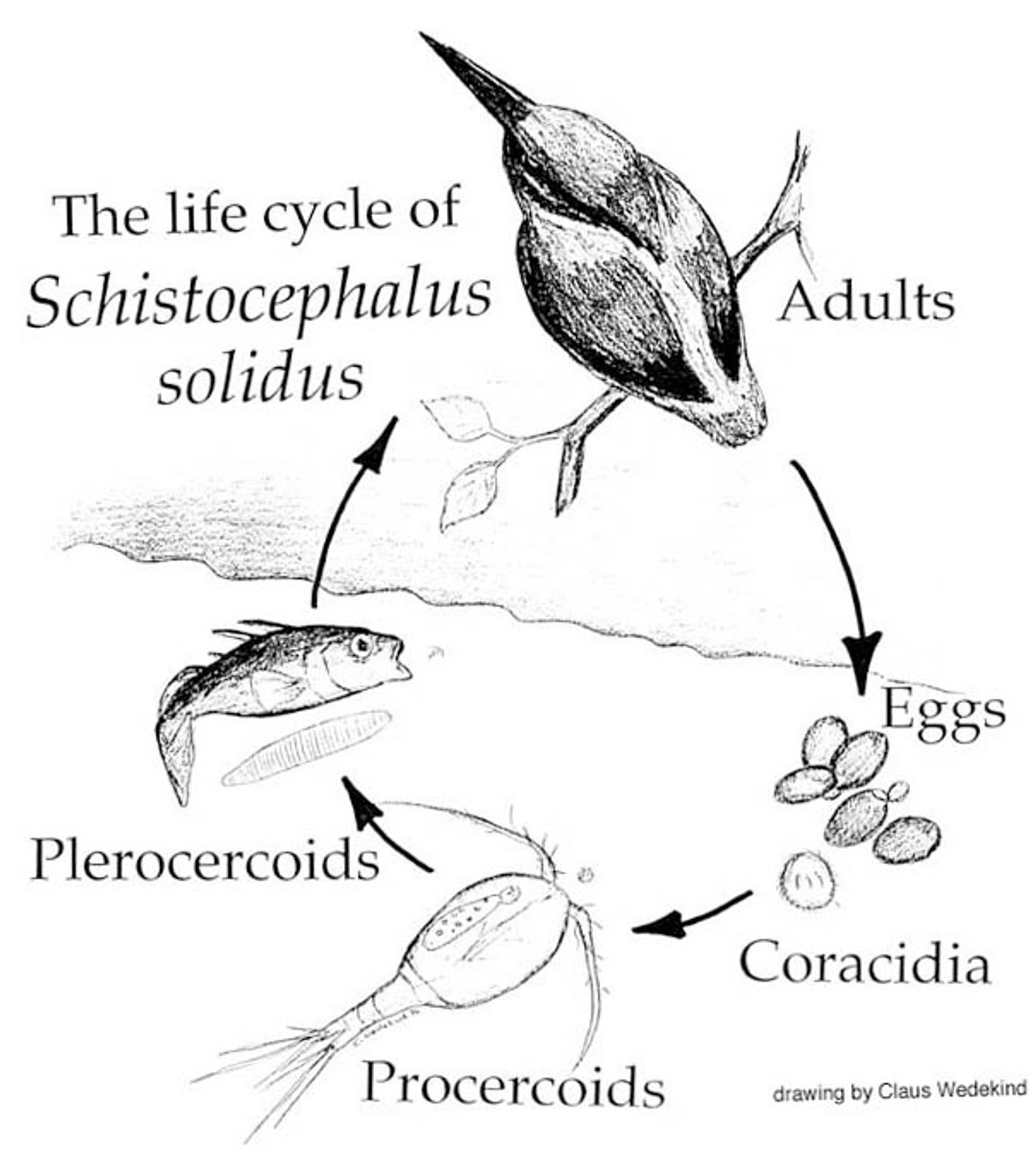
The molecular biology of Schistocephalus solidus, a parasitic tapeworm, was investigated by researchers at the Institut de Biologie Intégrative et des Systèmes and University of 22 Leicester in order to understand the life cycle of the microbe. S. solidus is a commonly used model in parasitology with a lot known about its physiology, yet very little is understood about the various genes that turn on and off throughout its incredibly complex life cycle.
This research paper aimed to characterize the transcriptome, or genes being ‘expressed’ as RNA, of the tapeworm during various stages of infection of a host. That meant an arduous process of sample collection under many different conditions, and a mountain of generated data to be analyzed.
Scientists from the Walter and Eliza Hall Institute of Medical Research and several other Australian institutions worked on the genetics of the human scabies parasite in the other
publication. Scabies has serious consequences for children living in Northern Australia, where scabies infections are common and can result in heart disease and seriously reduced life expectancy. Because of the microscopic size of the microbe - fractions of a millimeter, about one thousand mites must be pooled to get enough DNA to generate sequencing results.
Research papers often have limited space in which authors can describe what's been done, and often that means leaving out many crucial details about the methodology that others would likely need in order to replicate the work. These papers have been published in a way that allows readers to go directly to the complete, exact and detailed protocols used by the scientists themselves. It will also save time in the future because anyone who uses the ‘recipe’ will be able to simply cite what is already available online in future publications.
Because parasites live under such tremendously varied sets of conditions, parasitology is a complex field. The authors of these papers commented on how challenging it can be to perform good work that can be replicated, under these conditions. "Writing clear and accurate descriptions of the wet lab and bioinformatics methods is a challenge at the best of times. It is especially hard when the design is complex and requires iterative exploratory analysis using multiple tools. It necessitates great care and time consuming refinement of the text. I think documenting the methods using protocols.io will make this much easier," said Anthony Papenfuss, Associate Professor at Walter and Eliza Hall Institute and lead author of the scabies study.
First author of the tapeworm study François-Olivier Hébert explained that "We were able to [describe our complicated research] by making all of our homemade scripts, programs and datasets freely available to the public through GigaScience, GigaDB and protocols.io. They represent essential complementary platforms that allowed us to respect our vision of a reproducible science."
Sources:
AAAS,
GigaScience,
Nature