Rethinking the Way Viruses Are Classified
Scientists may have to start reclassifying viruses, according to new work reported in Nature Communications. The structure of a virus was thought to be critical to their classification, but in reality, they have many more patterns in their architecture than we knew. This work may also change our understanding of how viruses form, evolve, and cause infection. Scientists may now find new ways to target different viral shapes with vaccines.
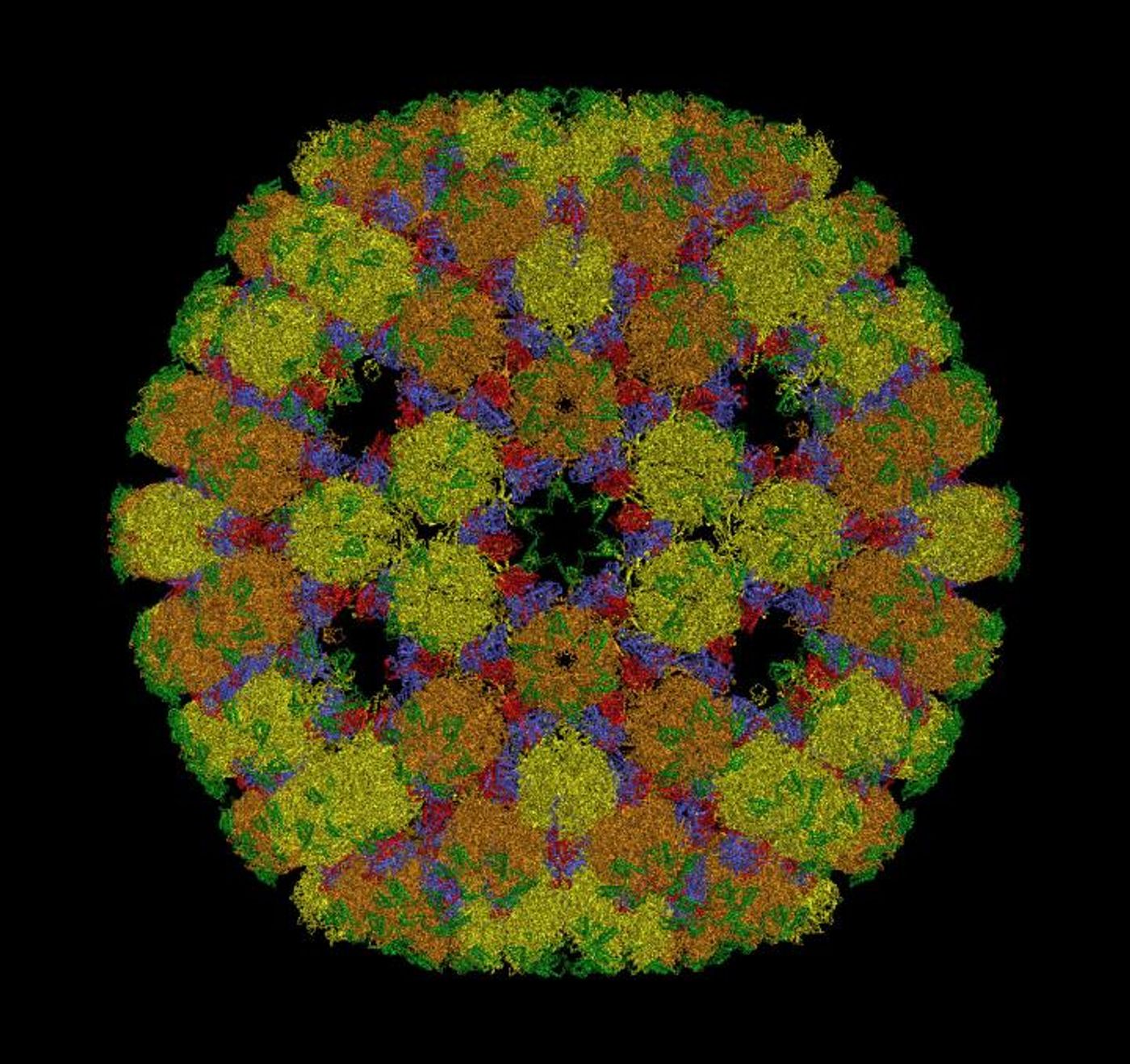
High-resolution images obtained in the 1950s and 60s revealed that viruses are contained within a protective cell called a capsid. Capsids are made of multiple copies of the same protein and were thought to have a shape like a 20-sided die; they were thought to have either helical or icosahedral shapes, for the most part. Capsids have different configurations to contain viral genomes of different sizes.
Now, theoretical biophysicist Antoni Luqueof San Diego University and mathematical biologist Reidun Twarock, of the University of York have found that while cryo-electron microscopy produced high-quality structural images of viruses, the math that described their architecture wasn’t outlined. Thus, many viruses have been misclassified, including commonly known viruses like Zika and Herpes simplex.
"We discovered six new ways in which proteins can organize to form icosahedral capsid shells," Luque said. "So, many viruses don't adopt only the two broadly understood capsid architectures. There are now at least eight ways in which their icosahedral capsids could be designed."
The researchers showed that viruses with the same structural lineage because their capsids are composed of the same protein take on a consistent icosahedral architecture. This information can now be used to classify viral structures in a different way, and reveal the evolutionary links between different viruses.
"We can use this discovery to target both the assembly and stability of the capsid, to either prevent the formation of the virus when it infects the host cell, or break it apart after it's formed," Luque said. "This could facilitate the characterization and identification of antiviral targets for viruses sharing the same icosahedral layout."
Viruses that were once outliers can now be accommodated by this classification system. Common geometric patterns shared by evolutionary distant but related viruses will now be identified and can help predict viral capsid architectures.
Twarock noted that these novel blueprints can provide "a new perspective on viral evolution, suggesting novel routes in which larger and more complex viruses may have evolved from simple ones at evolutionary timescales."
"This study introduces a more general framework than the classic Caspar-Klug construction. It is based on the conservation of the local vertices formed by the proteins that interact in the capsid," Luque explained. "This approach led to the discovery of six new types of icosahedral capsid layouts, while recovering the two classical layouts from Caspar-Klug based on Goldberg and geodesic polyhedra."
This work may also help inspire new ideas in architecture and design.
In these videos, different viral capsids are illustrated.
Sources: San Diego State University, AAAS/Eurekalert! via University of York, Nature Communications