The Genetics of Skin Inflammation, Seen With Unprecedented Clarity
A recent study published in Immunity details MIT scientists’ exploration of the underlying mechanisms of inflammatory skin conditions using a new RNA sequencing technology. The process of single-cell sequencing allows scientists to extract genetic information from individual cells using next-generation sequencing technologies. This technique not only allows researchers to examine extremely subtle differences between individual cells but also provides clarity around how cellular function is linked to its microenvironment.
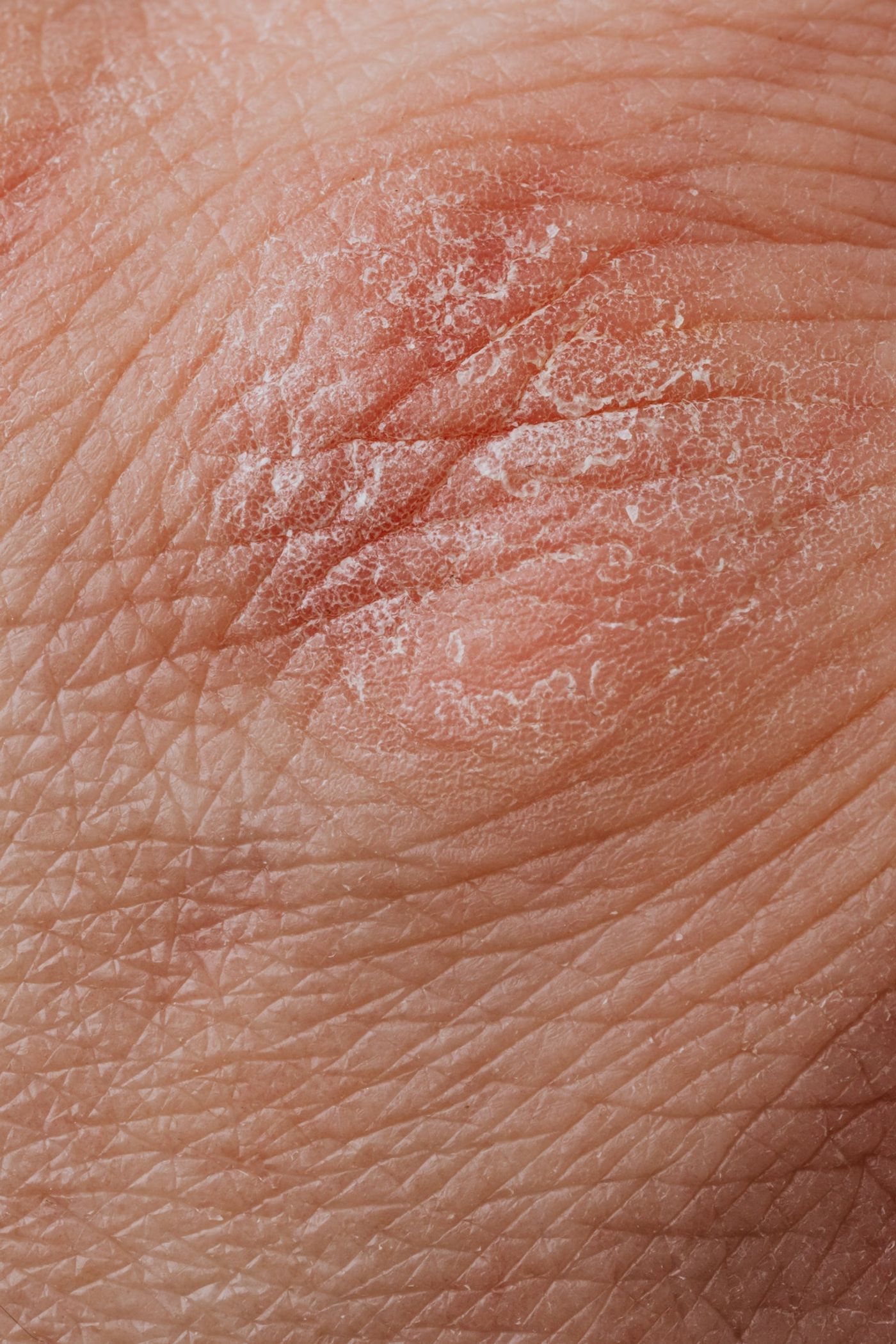
The team at MIT has turbocharged the resolution of the original RNA sequencing approach, known as Seq-Well. In the latest iteration of this technology, 10 times more genetic information can be extracted from each cell. Armed with this powerful methodology, the team sought to take a deep dive into the genetic landscape of five inflammatory skin diseases: psoriasis, acne, leprosy, alopecia areata, and granuloma annulare. Around 40,000 single cells were isolated from skin biopsies taken from patients with these various conditions.
“If you really want to resolve features that distinguish diseases, you need a higher level of resolution than what’s been possible,” explained J. Christopher Love, one of the researchers involved in the study.
“If you think of cells as packets of information, being able to measure that information more faithfully gives much better insights into what cell populations you might want to target for drug treatments, or, from a diagnostic standpoint, which ones you should monitor.”
Love and colleagues found signature patterns in the populations of T cells (immune cells involved in inflammation) that were activated in both leprosy and granuloma annulare, a chronic skin condition that causes ring-shaped lesions on the hands and feet. Additionally, in psoriasis patients, skin cells called keratinocytes that make up the outermost layer of the skin were found to be in a hyperactive state, dividing in overdrive.
According to study lead Alex K. Shalek, this sequencing technique opens up a whole new world of possibilities for disease research. “You never know what you’re going to want to use these datasets for, but there’s a tremendous opportunity in having measured everything,” said Shalek.
“In the future, when we need to repurpose them and think about particular surface receptors, ligands, proteases, or other genes, we will have all that information at our fingertips.”