Liver Let Die: Dying Liver Cells Disclose Cancer Diagnosis
Ranked as the third leading cause of cancer deaths worldwide, liver cancer is associated with a poor prognosis if caught in later stages. But long before liver disease develops into cancer, evidence of the impending disease is steadily released into the blood, only to disappear within hours.
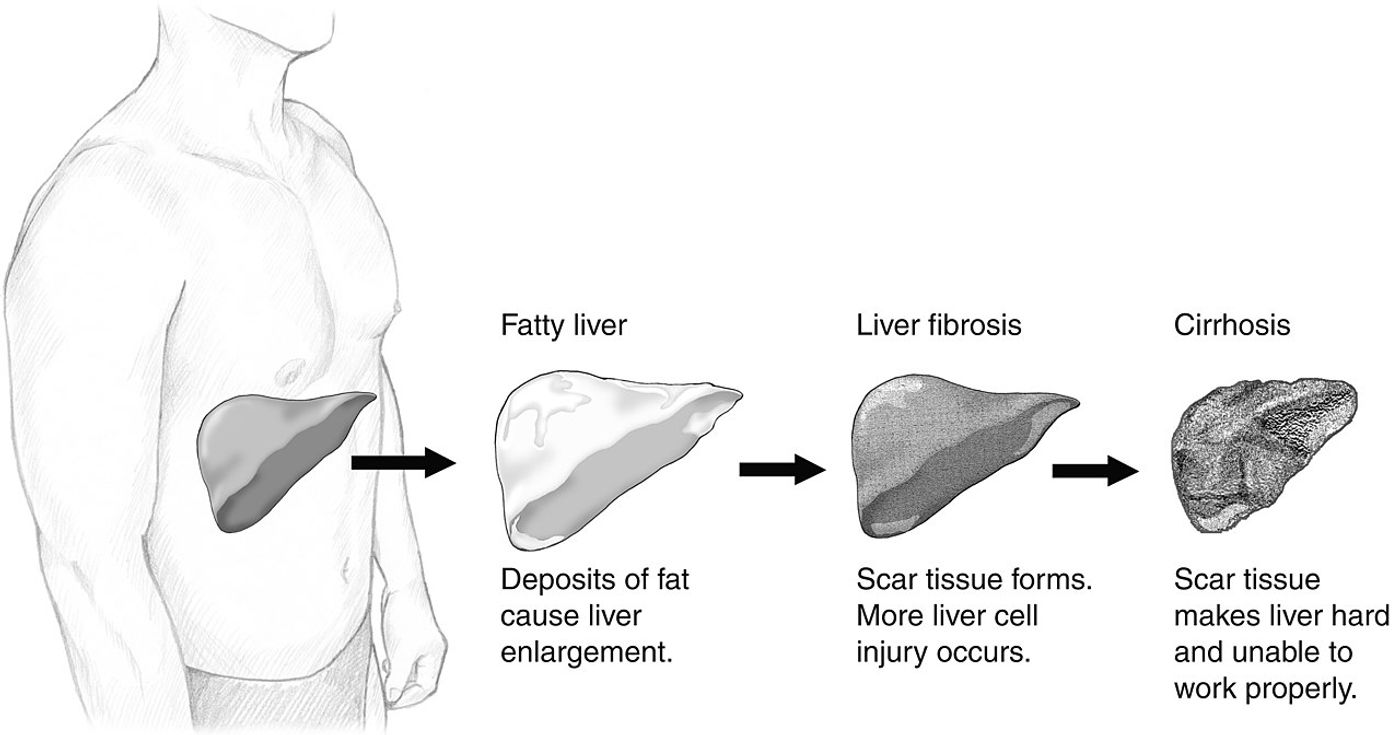
The stress of chronic liver inflammation due to chemical or genetic insults can lead to fibrosis, followed by cirrhosis. Irreversible, 10 to 20% of people with cirrhosis develop hepatocellular cancer (HCC).
Image Credit NIDDK, via Public Domain Mark 1.0
Before these dark times, liver tissue displays robust regenerative abilities. Up to 95% of liver cells are one of two different regenerating types. What type of liver cell you become depends on the transcription of specific regions of DNA. Methylation modifications to the DNA regulate cell-line fate. The epigenetic machinery regulating these modifications needs to work in perfect balance to expose only the right DNA for reading.
But obesity, hepatitis, and alcoholism insult these replicating cells, resetting their cell lineage programming back to a non-differentiated liver progenitor cell type. This creates a risk of the wrong type of liver cell growing in the wrong location.
Errant cells are often destroyed by necrosis or die via apoptosis, spilling their contents into the bloodstream. The gossamer threads of liver cell DNA linger for a couple of hours before being filtered away. This cell-free DNA (cfDNA) is dotted with specific methylation patterns. Like fingerprints, the patterns identify tumor-specific details and hint at what went wrong in the doomed cell.
This new and novel cancer biomarker is now the focus of a number of clinical trials. Several research companies are using liquid biopsies or blood draws to collect cfDNA from former liver cells to learn the different methylation patterns associated with various stages of cirrhosis and HCC.
At least four different groups pair cfDNA methylation panels with established diagnosis tools. The Mayo Clinic clinical trial collects liquid biopsies from liver disease patients with known diagnoses. Researchers hope to identify which methylation patterns are associated with which stage of liver disease.
One trial at the University of Taiwan pairs their blood panel diagnostic with whole genome sequencing to allow for finer distinction between the early stages of HCC. Another trial by Helio Genomics pairs a multi-analyte blood test with ultrasound diagnostic results.
But the front runners in this race towards a superior diagnostic is the German company Epigenomics, Inc paired with the USA-based Innovis LLC. In a 2021 study-related publication, they combine their previously developed HCC blood test with a next-generation sequencing (NGS) algorithm. Epigenomics Inc’s panel boasts high diagnostic levels of sensitivity (true positive detection) and specificity (true negative detection). The group confidently attributes the hypermethylation of the SEPT9 gene to abnormal cell division in liver carcinogenesis.
The identification of this new biomarker for liver cancer will lead to early detection, but these diagnostic applications are just the beginning. The truly crucial future step in this research is the development of therapies to prevent or reverse disease-state methylation.
Sources: Pharmacology & Therapeutics, Mayo Clinic, BioMed Central, New York State Department of Health