Favorable and Unfavorable Prognostic Genes in Human Cancers
Favorable and Unfavorable Prognostic Genes in Human Cancers
Cancer is a leading cause of death worldwide, so there is an urgent need to define the molecular mechanisms driving the development and progression of individual tumors and identify a molecular signature for cancer diagnosis and prognostic evaluation.
Patient treatment and prognosis are usually defined by cancer type, size, and stage at diagnosis, using, for example, the Tumor-Node-Metastasis (TNM) staging system classification.
However, these parameters alone do not accurately predict survival in patients because there are other clinicopathological factors and molecular markers that can determine prognostic estimation, such as specific genetic mutations and comorbidities.
The essential role of biomarkers in cancer research
Identifying cancer biomarkers by immunohistochemistry (IHC) in combination with primary antibodies is a widely used strategy in cancer diagnostics, screening, and research.
Tumor marker-specific antibodies can detect elevated quantities of tumor biomarkers in patients' blood, urine or tissue. Each tumor's biomarkers may be associated with more than one type of cancer.
A large number of molecular biomarkers, identified and validated in patients, are associated with the occurrence, progression, and prognosis of cancers. The majority of the biomarkers that have been extensively studied are proteins.
Biomarkers can be:
- Diagnostic: useful to determine the presence and type of cancer.
- Prognostic: give information on the patient's overall cancer outcome (favorable or unfavorable) with or without standard treatment.
- Predictive: provide information about the effect of therapeutic intervention, either in terms of tumor shrinkage or survival, compared to the patient's baseline condition (the concept supporting the "personalized medicine").
The difference between these biomarkers is clearly important, but often is misinterpreted or not recognized because many markers can be both predictive and prognostic. This misinterpretation could lead to delays in treatments and shorter patient survival times.
For example, a biomarker can be an unfavorable prognostic factor but could predict favorably for response to one therapy and unfavorably for another. Furthermore, a prognostic biomarker incorrectly labeled as predictive may result in overestimating the benefits of the treatment for a single patient or a subset of the population.
Prognostic biomarkers
Identifying prognostic markers aims to define patient subpopulations with significantly different anticipated outcomes which might benefit from various therapies.
Prognostic markers can be proteins, mRNAs, miRNAs, or the gene itself. For the latter, mutations, copy number aberrations, and single nucleotide variation (SNV) could also be prognostic.
Prognostic biomarkers give information on the patient's overall cancer outcome (favorable or unfavorable) with or without standard treatment.
- For favorable prognostic genes, higher relative expression levels at diagnosis give significantly higher overall survival for the patients.
- For unfavorable prognostic genes, higher relative expression levels at diagnosis give significantly lower overall survival for the patients
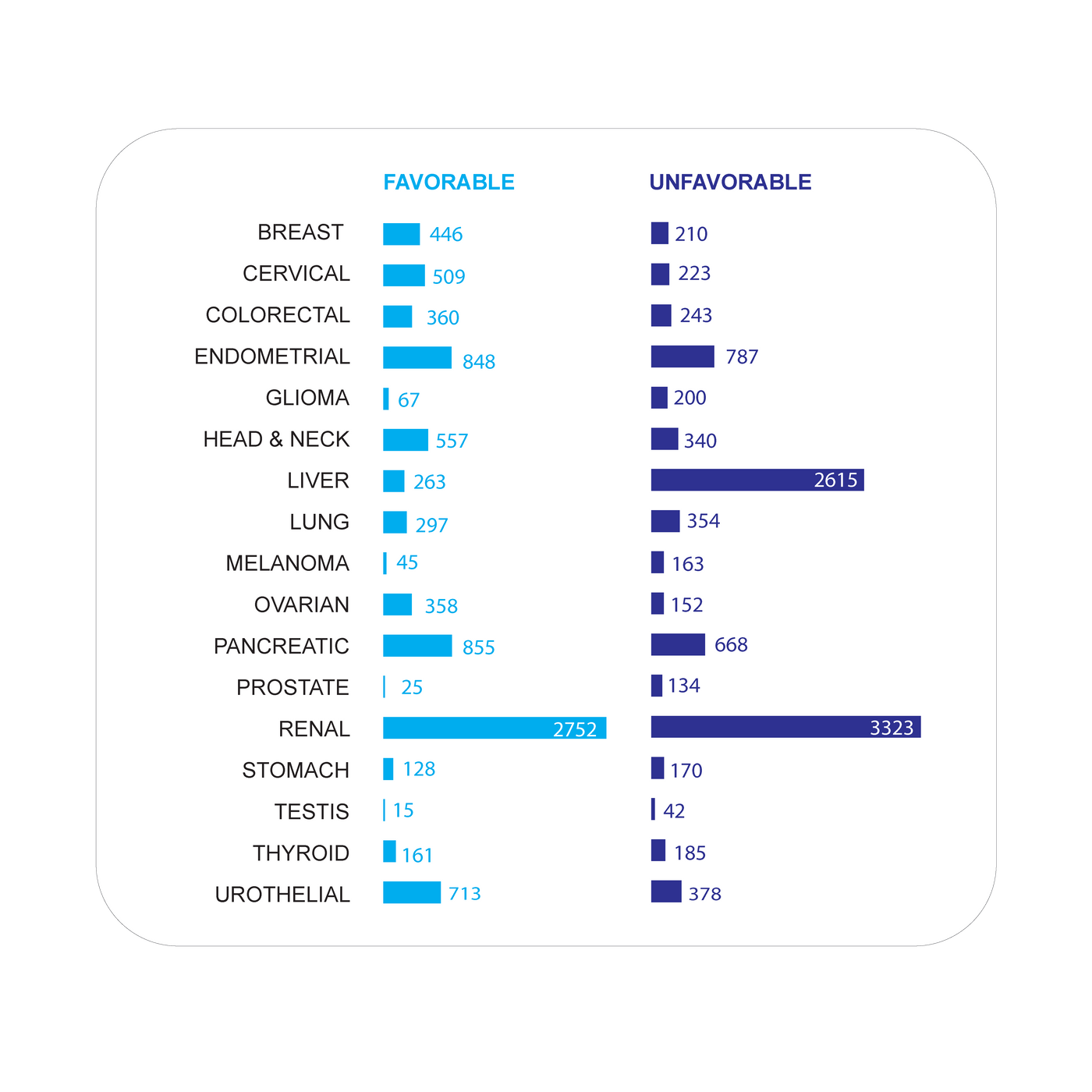
Figure 1. Favorable and Unfavorable Prognostic Genes in Human Cancers
The schematic summarizes the number of prognostic genes with regard to 17 cancer types and their association with favorable or unfavorable clinical outcomes. The classification is based on the Pathology Atlas, part of the Human Protein Atlas (proteinatlas.org).
Primary Antibodies for Cancer Research
The pathology Atlas to explore the expression profiles of human cancers
The Pathology Atlas is a section of the Human Protein Atlas containing information based on mRNA and protein expression data from 17 different forms of human cancer, together with millions of in-house generated immunohistochemically stained tissue sections images and more than 900,000 Kaplan-Meier plots showing the correlation between mRNA expression of each human protein gene and cancer patient survival.
This resource allows exploration of the specific genes influencing clinical outcome for major cancers, paving the way for further in-depth studies incorporating systems-level analyses of cancer. All data presented are available in an interactive open-access database to allow for genome-wide exploration of the impact of individual proteins on clinical outcome in major human cancers.
Explore the Expression Profiles of Human Cancers
Over 21,000 Triple A PolyclonalsTM from Atlas Antibodies helped to build the Pathology Atlas of the Human Protein Atlas.
Each antibody has been used for IHC-staining samples from 708 individuals originating from normal and cancerous human tissues. Trained pathologists have evaluated the reliability of all IHC-staining patterns.
Learn more about TripleA PolyclonalsTM
Example of Prognostic Biomarkers
Figure 2. Example of Unfavorable and Favorable Prognostic Genes.
In the Pathology Atlas a gene is considered prognostic if correlation analysis of gene expression and clinical outcome resulted in Kaplan-Meier plots with high significance (p<0.001). A gene may be prognostic in more than one cancer type.
A. JDP2 is an unfavorable prognostic gene in colorectal cancer. The patient's 5-year survival is 53% with high expression versus 64% for patients with low expression. The IHC staining using the Anti-JPD2 (HPA059511) polyclonal antibody, shows the differential expression pattern in colorectal cancer samples.
B. ABCD3 is a favorable prognostic gene in colorectal cancer. The patient's 5-year survival is 71% with high expression versus 36% for patients with low expression. The IHC staining using the Anti-ABCD3 (HPA032026) polyclonal antibody, shows the differential expression pattern in colorectal cancer samples.
C. REEP2 is an unfavorable prognostic gene in brain cancer. The patient's 3-year survival is 0% with high expression versus 10% for patients with low expression. The IHC staining using the Anti-REEP2 (HPA031813) polyclonal antibody, shows the differential expression pattern in glioma samples.
Download our brochure Atlas Antibodies in Colorectal Cancer Research
Explore primary antibodies targeting Glioma



![Everything You Need To Know About NGS [eBook]](https://d3bkbkx82g74b8.cloudfront.net/eyJidWNrZXQiOiJsYWJyb290cy1pbWFnZXMiLCJrZXkiOiJjb250ZW50X2FydGljbGVfcHJvZmlsZV9pbWFnZV9mNTM1ZjIyYzA5MDE5ZmNmMWU5NmI0ZDc4NWU2MzdiZTZlN2I5ZDk5XzE4NDUuanBnIiwiZWRpdHMiOnsidG9Gb3JtYXQiOiJqcGciLCJyZXNpemUiOnsid2lkdGgiOjcwMCwiaGVpZ2h0IjozNTAsImZpdCI6ImNvdmVyIiwicG9zaXRpb24iOiJjZW50ZXIiLCJiYWNrZ3JvdW5kIjoiI2ZmZiJ9LCJmbGF0dGVuIjp7ImJhY2tncm91bmQiOiIjZmZmIn19fQ==)