Tracing the Cells of an Animal to Their Origins
The many diverse parts of organisms are made up of different cell types, which have their own functions and characteristics. Relatively recent innovations have opened up single cells for analysis, and new work has investigated the ancestry behind individual cell types. Scientists have a host of questions they want to pursue.
"Whenever we use such a technology to examine an organ or an organism, we find not only familiar cell types, but also unknown and rare ones," noted Dr. Jan Philipp Junker, head of the Quantitative Developmental Biology research group at the Max Delbrück Center for Molecular Medicine in the Helmholtz Association (MDC). "The next question is obvious: Where do these different types come from?" Junker's lab has published a report in Nature Genetics detailing a technique called LINNAEUS; it allows scientists to learn the lineage and type of every cell in an organism.
"We want to understand the flexibility with which organisms develop," Junker said.
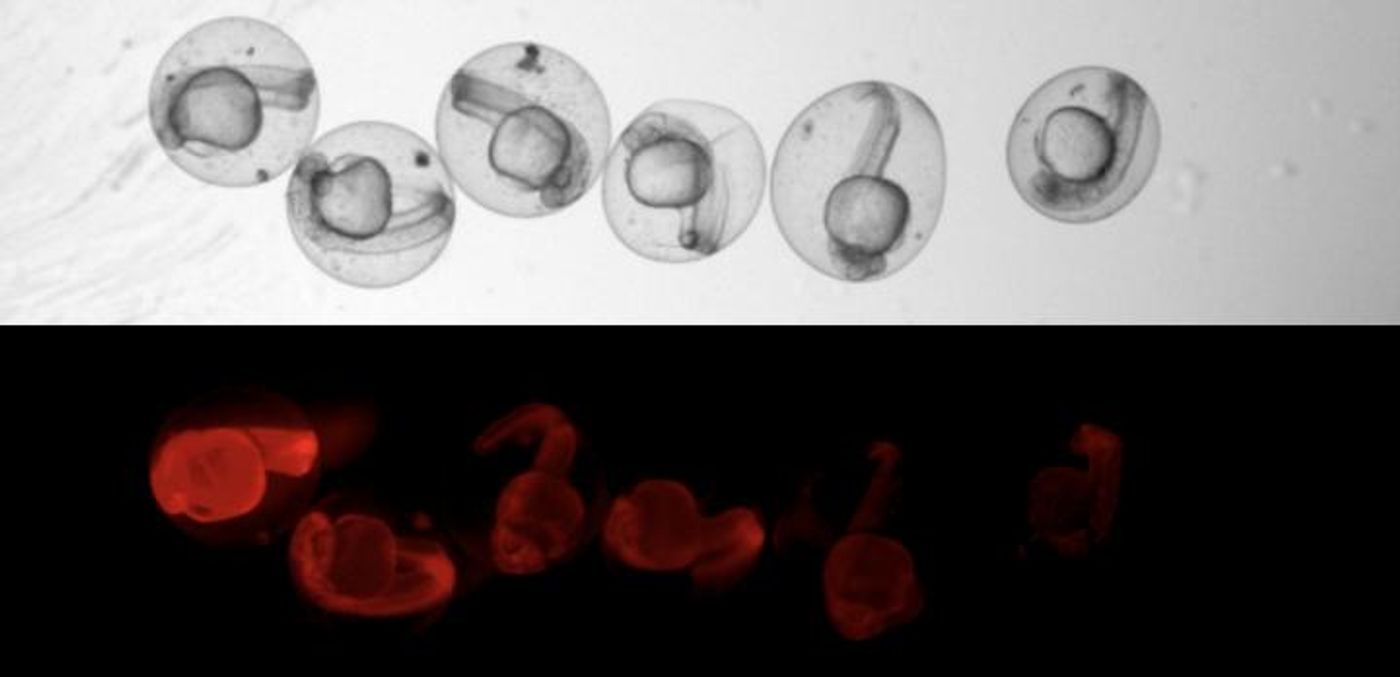
LINNAEUS uses scars in DNA, which act as a kind of barcode that allows the cells to be traced through their development. The scars are generated with the CRISPR/Cas9 gene-editing system (outlined in the video), which in this case is targeting a gene for red fluorescent protein (RFP), so the edits are harmless to the fish and they develop normally.
"CRISPR always makes a cut at an exact point. But the cells have no more than 15 minutes to repair before the next cell division takes place," Junker explained. "Patching must be done quickly, so the chromosome pieces are sort of glued together. This is where mistakes happen. The scars in the DNA are of random length and their exact position also varies." After scarring, any daughter cells inherit the scar, so cells with the same ancestor are easy to identify.
The investigators had to account for some of the difficulties inherent in their process. The massive amount of data was sure to include erroneous sequences. “If the same scar sequence is created in both heart and brain cells, one might mistakenly assume that they have a common ancestor," Junker says. So we had to know which sequences we couldn't trust and filter them out." Additionally, not all scars in a cell can be found, said a lead author of the study, bioinformatician Bastiaan Spanjaard. "So we have developed a method capable of bridging gaps in the data that enables us to construct lineage trees."
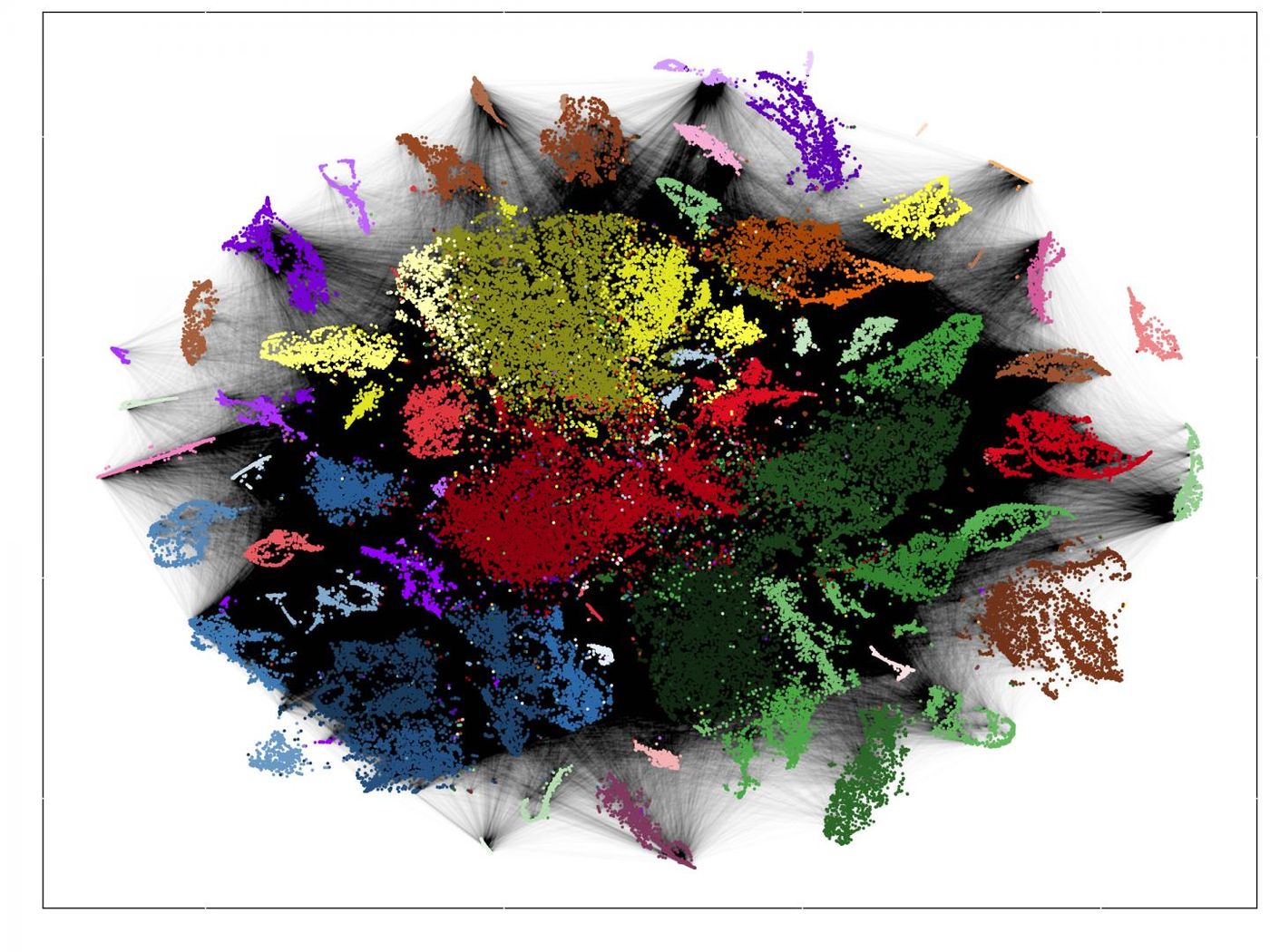
The analysis produced colorful charts that show how cells branch off, each color representing a different cell type. The large dataset can be seen in detail by zooming in.
"In the heart, for instance, there are two cell types that are barely distinguishable. But the lineage trees show that their development branches off in different directions very early on," said Junker. "We next want to see where these cell types occur in fish hearts. This often provides a first indication of what function they serve."
The researchers in Junker’s lab use zebrafish, but they may extend their studies to human organoids. This work will help us understand the effect of mutations that occur at different times or places.
Sources: AAAS/Eurekalert! Via MDC, Nature Biotechnnology