Geneticists are learning not to dismiss something just because they do not understand it. In the realm of studying non-coding RNAs (ncRNAs), a new technology presented by researchers from the University of Toronto just changed everything.
Image Credit: www.wired.co.uk
So-called “junk DNA” makes up practically 98 percent of the 3 billion strong population of nucleotide letters in the human genome, yet they do not code for proteins like the other 2 percent. Scientists used to believe that DNA sequences not coding for proteins were simply a waste of space, but these sequences may actually play a significant role in disease progression, as 50 to 75 percent of them are transcribed into RNA like their protein-coding cousins, making ncRNAs.
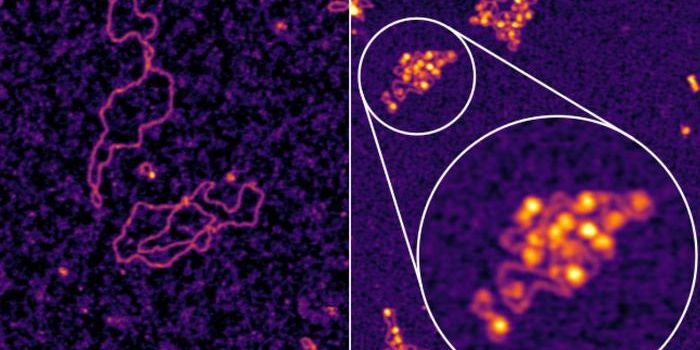
Scientists have not been able to fully understand the role of ncRNAs because of a lack in technology for visualizing their activity. With the development of a technique called “Ligation of Interacting RNA followed by high-throughput sequencing” (LIGR-Seq), University of Toronto scientists are shining a light into the dark tunnel of ncRNA activity.
"Up until now, with existing methods, you had to know what you are looking for because they all require you to have some information about the RNA of interest,” said Eesha Sharma, PhD candidate. “The power of our method is that you don't need to preselect your candidates, you can see what's occurring globally in cells, and use that information to look at interesting things we have not seen before and how they are affecting biology.”
LIGR-Seq measures and analyzes activity and interaction between different RNA molecules by pairing RNAs with matching sequences together, extracting them from live cells, and identifying the specific type of ncRNA and their potential functions. There are more than seven types of ncRNAs.
What is it that ncRNAs do? Scientists used to think that there existence with arbitrary, but some believe that they play a small role in gene regulation. Some types of ncRNAs have been discovered to act as “carriages for shuttling mRNAs around the cell,” providing assistive functions during the transition from RNA to protein. With the invention and implementation of LIGR-Seq, scientists can observe these functions and more, potentially uncovering the role of ncRNAs in cancer metastasis.
LIGR-Seq provides a way for scientists to complete a “systematic functional analysis” of ncRNAs, and researchers from the University of Toronto have already made their own discoveries about a new role of a particular type of ncRNA, called small nucleolar RNA (snoRNA).
They knew already that snoRNAs facilitate the chemical modifications of other ncRNAs, and further LIGR-Seq experiments showed that snoRNAs also maintain the stability of certain protein-coding messenger RNAs, meaning they are able to influence the types and amounts of proteins made.
Now capable of discovering new RNA pairs and understanding their interactions at an unprecedented level with LIGR-Seq, University of Toronto scientists have many plans for future studies. They predict investigating how ncRNAs function during development, their role in neuron formation, and their involvement and interaction with each other in disease progression.
The study was recently published in
Molecular Cell.
Source:
University of Toronto