When we think of genetic mutations we usually think of the mutations that cause disease, but protective mutations are an under-researched area. It can be very difficult to identify a disease-causing mutation and it is even more difficult to try and identify a protective mutation, as demonstrated by a recent paper from senior authors Eric Schadt and Stephen Friend from the Icahn School of Medicine at Mount Sinai. The title of this paper, published in Nature Biotechnology, does a good job of summarizing what they accomplished: “Analysis of 589,306 genomes identifies individuals resilient to severe Mendelian childhood diseases.” From their analysis of over half a million genomes, they found 13 people that, according to their genetics, should have had a debilitating childhood illness, but somehow are perfectly healthy.
By uncovering any potential protective mutations for Mendelian diseases, it might be possible to discover new mechanisms of disease and new therapeutic targets. Developing treatments for Mendelian diseases is surprisingly difficult for a number of reasons. A big reason for the lack of biological cures is that a lot of the mutations are loss of function (LoF) mutations, and that is a difficult problem to fix with a therapeutic, It is nearly impossible to restore a protein’s function with a small molecule. While small molecules aren’t the only option for therapeutics, they are generally the least expensive and most convenient. Because of these issues with developing a full cure, most treatments for these types of disorders are focused on symptom management.
Looking for protective mutations isn’t really that much of an out-of-the-box idea; there are many well-documented protective mutations. For example, a rare mutation in
CCR5 confers resilience against HIV infection. There are also documented LoF mutations in
PCSK9 and zinc transporter 8 that protect against heart disease and diabetes, respectively. This comprehensive study identified 13 people with protective mutations against 8 different diseases: cystic fibrosis, Smith-Lemli-Opitz syndrome, familial dysautonomia, epidermolysis bullosa simplex, Pfeiffer syndrome, autoimmune polyendocrinopathy syndrome, acampomelic campomelic dysplasia and atelosteogenesis. While the researchers know that these people have a mutation that would lead to one of these diseases, the protective mutation is impossible to elucidate with the amount of data the researchers have access to. Unfortunately, for most of the over half a million genomes that they had access to, they only had access to the data, with no possibility of re-contact with the person. With more access to the 13 people with protective mutations, the likelihood of figuring out the protective mechanism would be much higher.
Another issue is that analysis of 589,306 genomes only produced 13 people. Are protective mutations so rare? The small amount of people with resilience to severe Mendelian diseases is more of a function of the stringency filters the researchers used in their analysis. They performed another analysis with slightly less strict filters to produce another 111 people with suspected resilience. However, by loosening the filters, the analysis of who actually has resilience becomes much more complex. Instead of actual bona-fide resilience, it could be that that particular Mendelian disease is not as penetrant as they thought. Another issue that the researchers ran into was misdiagnosis of the resilient patients because their symptoms were not canonical or were perceived as sub-clinical. Keeping the stringent filters, while possibly overlooking some resilient individuals, greatly increased the confidence in the patients that it did produce.
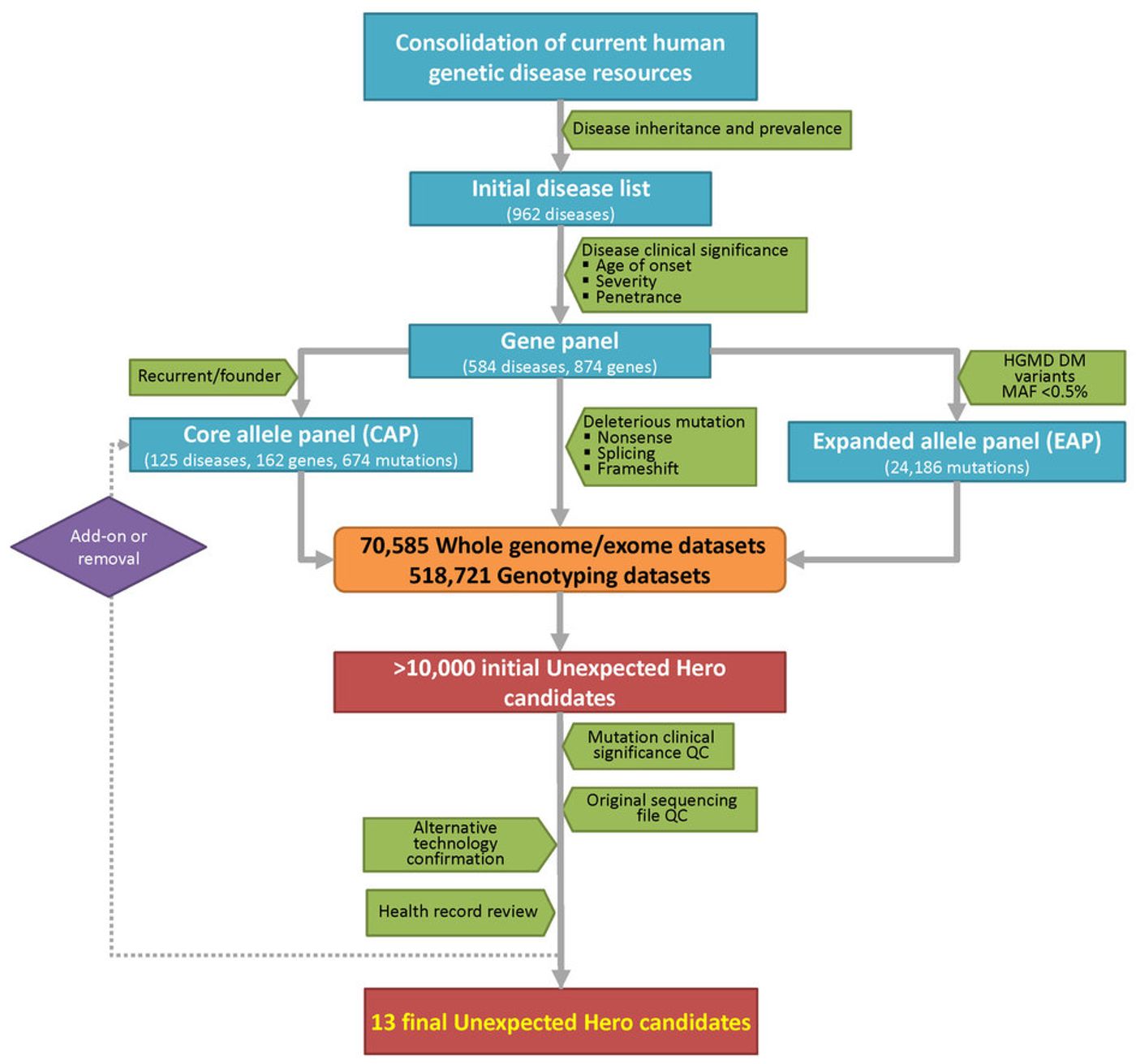
Genetic resilience analysis workflow.
Image Credit: Chen et al. Nature Biotechnology, 2016.
I never really thought about looking for individuals resilient to disease as a way to figure the mechanisms of disease or to identify new therapeutic targets, but this paper opened my eyes to the utility of this kind of approach. This paper is also a study in using readily available big data to pull out novel and useful information. While the analysis could still be refined, this is a great start to an interesting field.
Sources:
Nature Biotechnology and
AlzForum