A Major Step Toward a Vaccine for Severe Malaria
The World Health Organization estimates that there were about 219 million cases of malaria in 2017, which caused around 435,000 deaths. Scientists have now learned more about malaria proteins and the antibodies that combat them, which gets us closer to creating a malaria vaccine against the worst forms. The findings have been reported in Cell Host & Microbe.
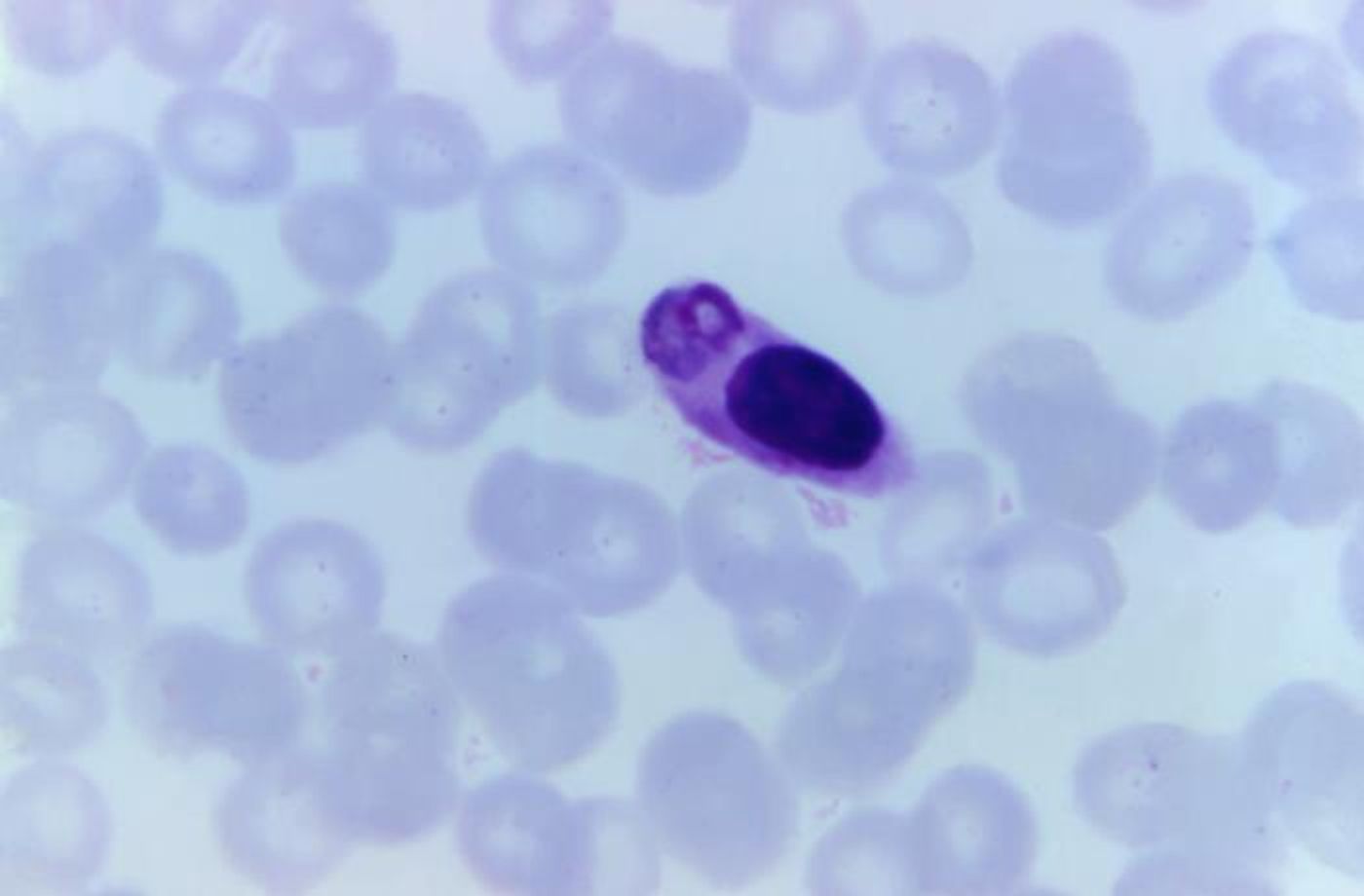
Malaria parasites have a complex life cycle, and when they reach a human host, they head for the liver. They multiply there and are then released into the bloodstream, where they infect red blood cells and destroy them. Associate Professor Alyssa Barry, leader of the Systems Epidemiology of Infection at Deakin University, explained that once the parasites get into red blood cells, they add proteins called PfEMP1 to the surface of these cells.
"As part of their survival strategy within the human host, malaria parasites use PfEMP1 to stick to the walls of blood vessels, and this can cause blockages to blood flow and inflammation, leading to severe disease," Barry said. "Malaria parasites change these proteins to escape from developing immune responses, and every strain has a different set of proteins, making the identification of vaccine targets like finding a needle in a haystack."
By studying hundreds of variants of PfEMP1 proteins, a team of researchers has identified the antibodies that are best at battling severe forms of malaria. The scientists gathered hundreds of PfEMP1 proteins isolated from children in Papua New Guinea (PNG) that had been naturally infected with various strains of malaria.
"It's the first time anyone has shown this. For years, researchers have thought that developing a malaria vaccine based on PfEMP1 would be virtually impossible because the proteins are just so diverse," noted Barry.
"It's similar to the flu vaccine, where you have to keep adjusting and updating it as the virus strains evolve from year to year. Malaria is even more diverse than influenza; one village in a country such as PNG could contain thousands of possible PfEMP1 variants," Barry continued. "But in malaria-endemic areas, children who are repeatedly infected develop immunity to severe malaria by the time they're about two years old, so we know antimalarial immunity is possible, and it can develop after exposure to only a few strains."
Immunity to mild forms of the diseases was an obstacle to the work, but when malaria becomes severe, the immune system only targets a small group of proteins that are similar in different strains. This makes it easier to create a vaccine that would target these shared proteins.
Learn more about the lifecycle of the malaria parasite from the video.
"Using genomic sequencing, we collected PfEMP1 proteins from different strains of malaria, measured antibodies to those proteins to identify the protective antibody - the biomarker of immunity - that protects kids against disease," Barry added. "We were able to identify these antibodies by monitoring for patterns of disease, following the children in PNG for 16 months to determine which of them were susceptible to the more severe forms of the disease, and those who were protected and only experienced milder forms of the disease.
"Our study shows the potential of new approaches that combine measurement of large numbers of antibodies and carefully designed epidemiological studies," said Dr. Sofonias K. Tessema, who began the work as Barry's student and is now a postdoctoral fellow at University of California San Francisco.
Sources: AAAS/Eurekalert! via Deakin University, Cell Host & Microbe