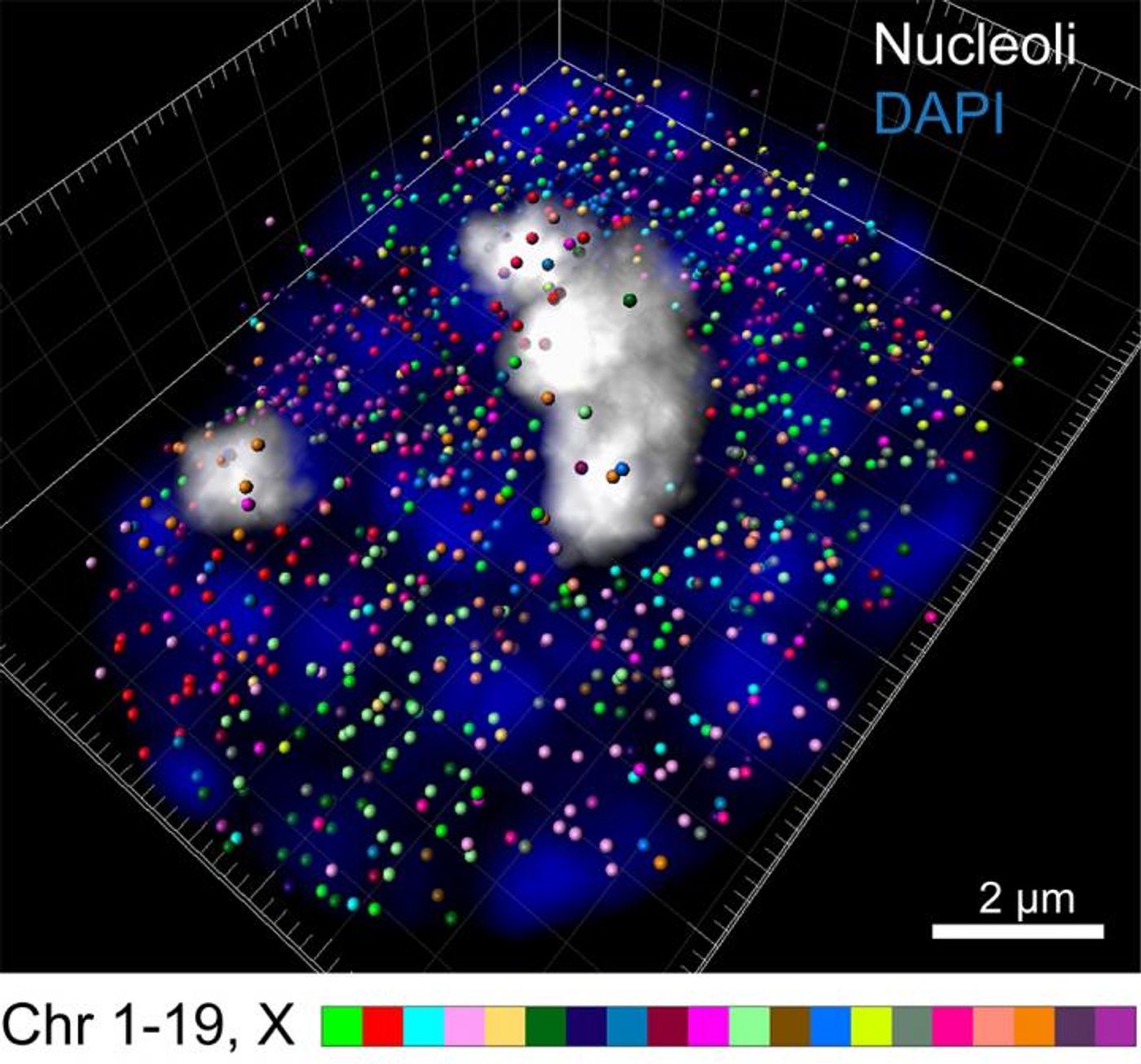
With labeling tools that add fluorescent tags, researchers could once see about four or five genes in a cell. Updates to that technique by scientists at Caltech will now enable the visualization of over 10,000 genes in one cell at the same time. This new method has been called intron seqFISH (sequential fluorescence in situ hybridization) and has been reported in the journal Cell.
Long Cai, research professor in biology and an affiliated faculty member of the Tianqiao and Chrissy Chen Institute for Neuroscience at Caltech led this study, which utilized introns. When genes in DNA are active, they are transcribed into RNA, an intermediary molecule the cell usually uses to make proteins. Before being made into protein, the RNA has to be modified; one edit is the removal of introns, which are non-coding regions of DNA that sit in between exons, the coding portions of genes. The investigators labeled introns because they are only around when transcription first starts. Thus, they could get a snapshot of gene activity at one moment.
In the intron seqFISH method, every intron gets a fluorescent label, which can be seen under a microscope. That indicates which genes are active and can also show how high their expression level is. In all, the researchers barcoded and visualized 10,421 genes at one time.
Previous barcoding tools have used RNA, which can show how the activity of genes changes over time. The intron labels highlighted the newly transcribed genes, the so-called nascent transcriptome. When they used the tool, the investigators were surprised by what they found; they saw that genes are transcribed on a short, oscillating course, with many genes bursting on and off over two hours.
These oscillations haven’t been observed before because of technical limitations. Intron seqFISH allowed the scientists to actually see what they measured in a single cell, ensuring it was not an artifact. Introns also don’t hang out for as long as mRNA does, so it’s easier to see them coming and going.
Introns tend to stick around the gene, so the labels also allowed the investigators to see the locations of genes in chromosomes, where the entire genome is compacted in the nucleus of cells. They saw that genes encoding for protein aren’t buried within the chromosome but are instead near the surface.
"This technique can be applied to any tissue," said Cai. "Intron seqFISH can help identify cell types and also what the cells are going to do, in addition to giving us a look at the chromosome structure in the same cells."
Learn more about the fluorescent in situ hybridization technique from the video.
Sources: AAAS/Eurekalert! Via Caltech, Cell