Digital PCR (dPCR) in oncology applications: when accurate counts really matter
When it comes to detecting rare cancer-causing mutations, both sensitivity and specificity matter. In the past few years, digital PCR (dPCR) has captured the attention of researchers and clinicians alike as an up-and-coming technology to complement existing methods for detecting cancer cells and cancer-associated genome variants—especially in applications where sensitivity is especially important, such as liquid biopsy-based disease monitoring or the detection of copy number alterations (CNAs).
Like other PCR workflows, dPCR employs primers flanking the target regions, a dye or fluorophore to label the amplification products, and a method to detect the signal following amplification. However, dPCR differs from these other methods in several important ways.
- First, the reaction mixture is divided into thousands of tiny microreactors either by separating the reaction into nanowells on a chip or plate, or by creating emulsions of tiny “reactor droplets”; this isolates rare target molecules from potential background “noise” so they are easier to detect.
- Then, following amplification in the presence of fluorophores, each microreaction is counted as either positive (1) or negative (0) for the target molecule, and Poisson analysis determines the number of target molecules in the original input.
- Finally, dPCR provides sensitive, accurate quantification of target molecules without requiring a set of standards, enabling the detection of rare molecules or even small changes in copy number.
Identifying copy number alterations (CNA) with dPCR
Copy number alterations are regions of the genome that have been deleted or duplicated, leading to either too few or too many copies of genomic elements. CNAs (also referred to as CNVs, or copy number variations) vary widely in size, from just a few nucleotides to entire chromosomes (as in trisomy 21, or Down syndrome). CNAs are known to contribute to many complex human diseases, including neurological diseases and many cancers—such as breast, lung, prostate, and colorectal cancers. The ability to accurately quantify these CNAs can be critical to understanding specific risks, prognosis, and treatment options.
Digital PCR outperforms many established methods for detecting CNAs. For example, in addition to having a short turnaround time, the high quantitative and qualitative accuracy of dPCR is superior to methods such as: qPCR, which requires standards and also provides low resolution, leaving it susceptible to false negatives; DNA microarrays, which are costly, require more input sample, and offer low resolution; FISH (fluorescent in situ hybridization), which is expensive, time-consuming, and low-throughput; and next-generation sequencing (NGS), which is costly, complex in terms of workflow and analysis, and prone to variable false-positive rates.
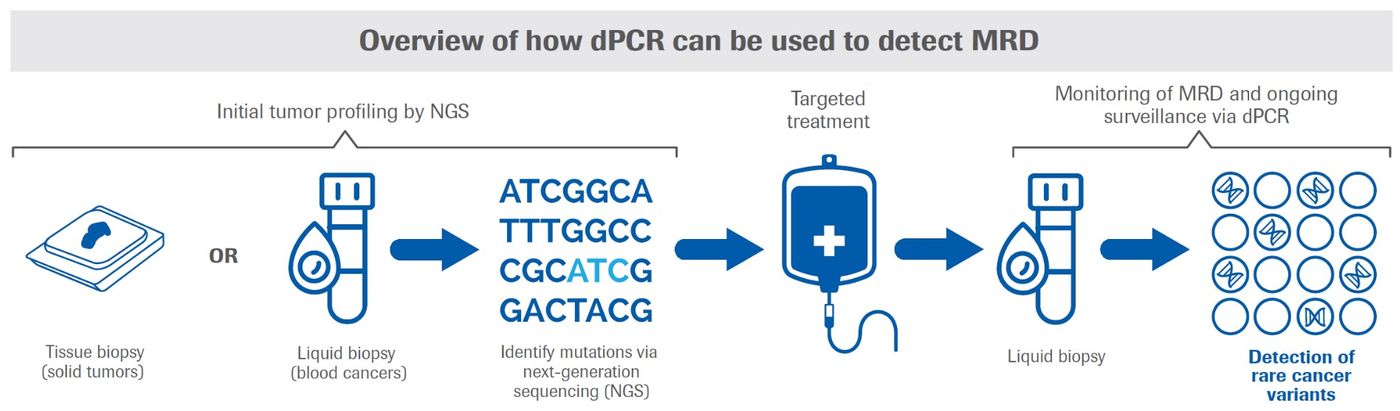
Another oncology application in which dPCR shines is the detection of very rare mutations or cancer cells, such as those found in MRD (now also described as “measurable” residual disease). MRD refers to rare circulating tumor cells that may remain following treatment for cancers such as lymphoma, multiple myeloma, acute lymphocytic leukemia, and some solid tumors (see Figure). While the number of residual cancer cells may be very small, identifying them is important because they may have the potential to expand, causing relapse.
For solid tumors, dPCR can detect ultra-rare tumor cells in liquid biopsy samples with at least 10-fold greater sensitivity than either qPCR and NGS; in practical terms, this means a much lower likelihood of missing remaining cancer cells that could lead to a relapse. dPCR is also more sensitive and less time-consuming compared to other methods for detecting MRD, such as flow cytometry and the microscopic examination of bone marrow samples.
In summary, dPCR offers a highly sensitive method for accurate quantification of target mutations in both noninvasive liquid biopsy samples and solid tissue samples, using methods that are more sensitive, faster, more reliable, and less complex than many other workflows.