A Better way to Develop new Drugs
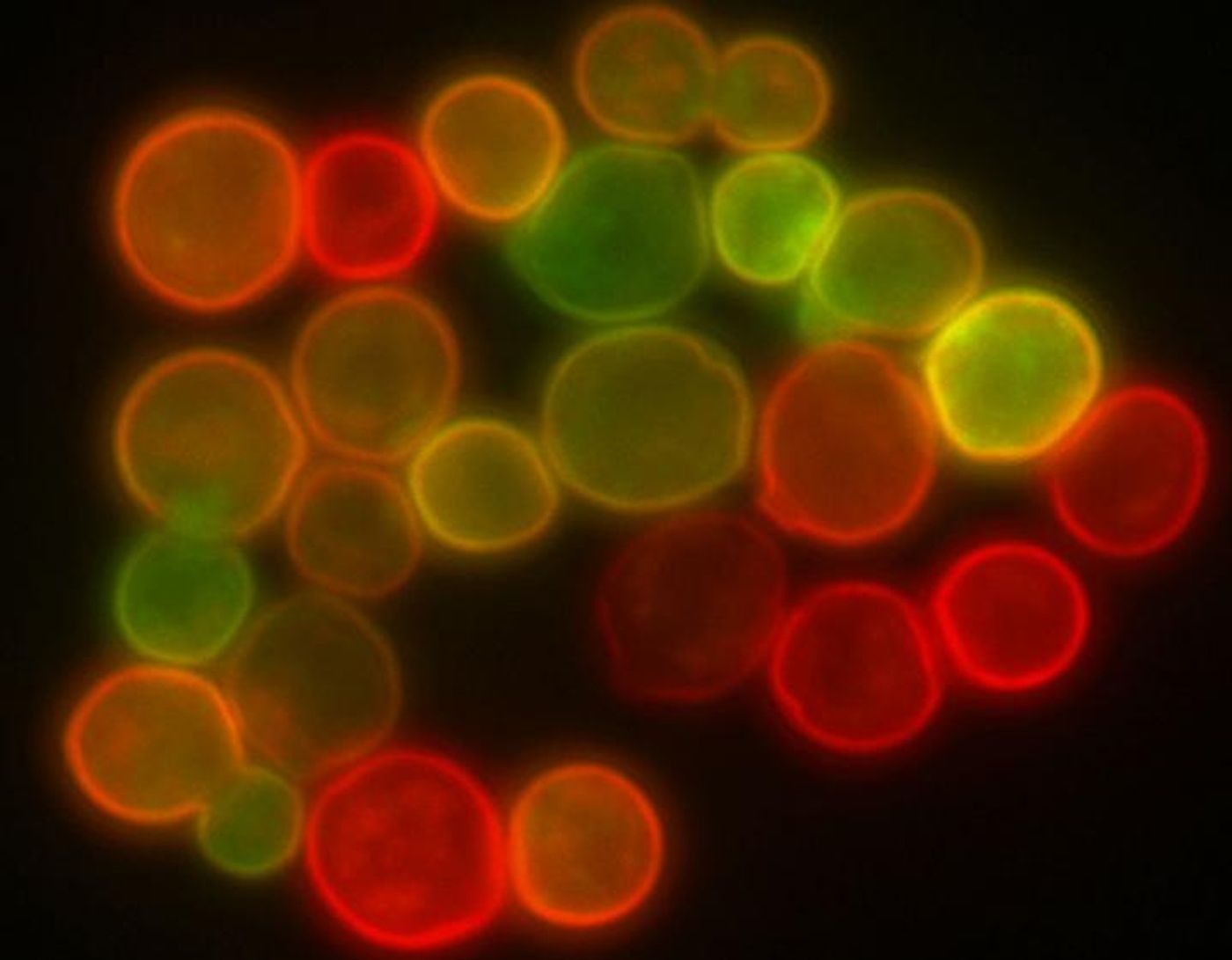
Finding new drugs can be challenging. While researchers might gain a solid understanding of a biochemical pathway or drug target, it may be tough to find a compound that is suitable for a specific need. The other option is to generate new compounds at random, and then see what they do when tested. To try to develop a better approach, researchers have used chemical genetics in a yeast model to learn more about how drugs act on cellular pathways.
This new work assessed the cellular impact of around 14,000 compounds, including hundreds of novel molecules, in an attempt to spread the word about chemicals that have potential as disease therapeutics. The study, which was reported in Nature Chemical Biology, identified found 1000 candidates for treating disorders ranging from cancer to Alzheimer’s.
Many of these new compounds were derived from microbes that live in soil, one place where scientists from several groups are currently looking for new drugs.
Drug discovery is still largely a game of guessing and luck, where random libraries of compounds are screened against targets in a search for matches. “There are many different types of libraries to choose from. A lot of the time you choose a library based on its availability or its cost, not any sort of functional information, and so it becomes a shot in the dark," explained lead author Dr. Jeff Piotrowski, formerly a postdoctoral fellow in both the Yoshida and Boone labs. He is now with Yumanity Therapeutics.
Related: New, Powerful Antibiotic Found in Soil
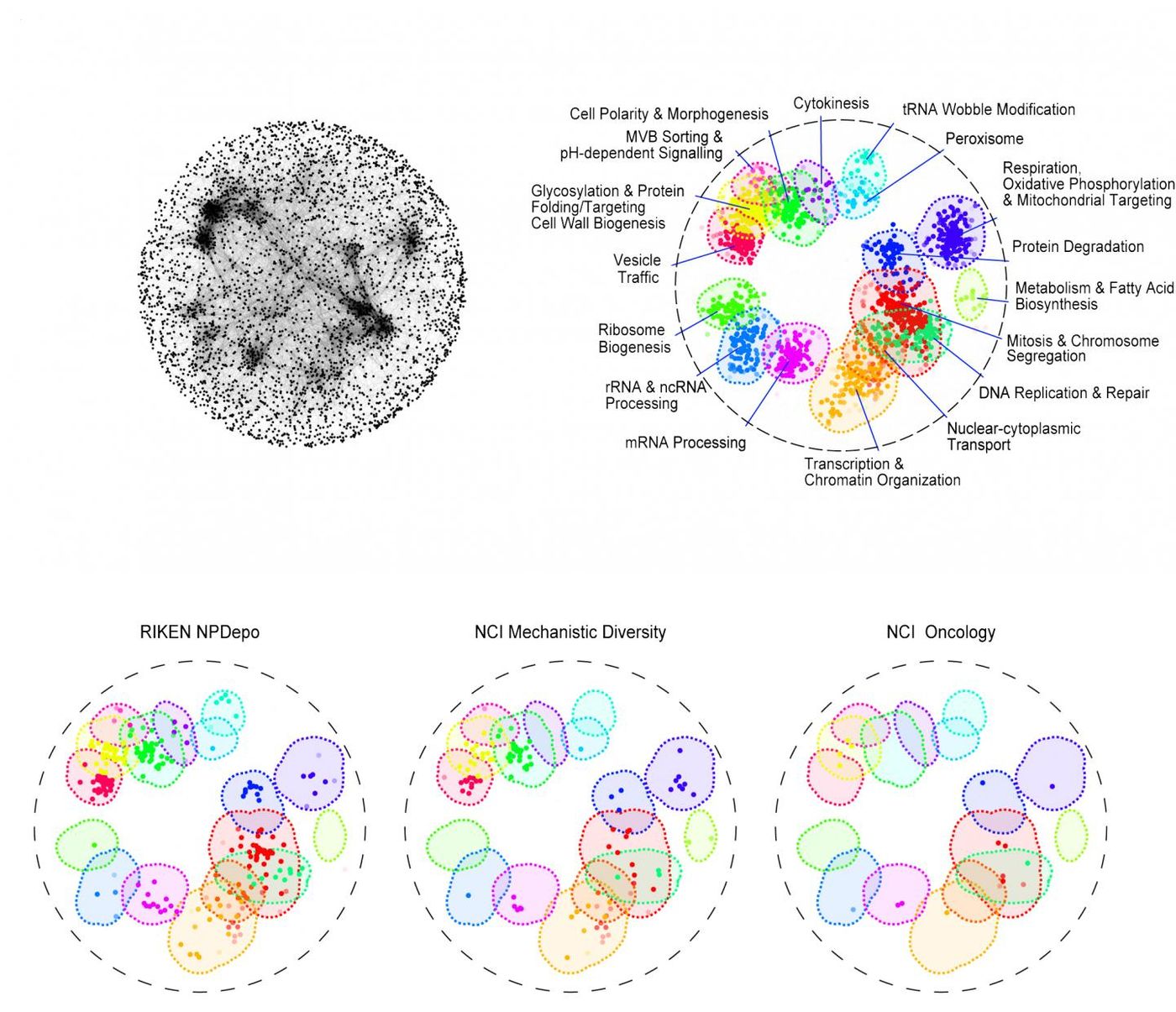
Piotrowski and colleagues used a high-throughput method to demonstrate the cellular targets of thousands of molecules from seven libraries, including libraries from the National Cancer Institute (NCI), the National Institute of Health, Glaxo-Smith-Kline and from RIKEN in Japan; the RIKEN group contains thousands of untested compounds made by soil microbes.
"By annotating these libraries, we can tell which library targets which bioprocess in the cell. It gives us a head start on linking a compound to a target, which is perhaps the most challenging part of drug discovery," said Piotrowski.
The RIKEN library turned out to be one of the richest. Some of those compounds have the potential to fight infections; others could interfere with processes implicated in Alzheimer’s and Parkinson’s. Many of the RIKEN compounds show promise as cancer drugs as well.
These compounds are produced by soil microbes, and as natural chemicals would likely be better at entering cells. Many of our most important medicines are products of the natural world, like penicillin or taxol.
"It's long been thought that natural products are more functionally diverse, that they can do more things than purely synthetized compounds and that certainly seems to be true from our data," explained Charles Boone, a Professor of Molecular Genetics in the University of Toronto's Donnelly Centre.
Because so much is known about how biological processes work in yeast, they were a perfect model for this study. Researchers used previous work by Boone, Myers, and Donnelly Centre Director Brenda Andrews that investigated how gene interactions drive basic cellular functions. The idea is that there may be compensation for the loss of a single gene, but the loss of two genes causes a major impact, one that reveals the functionality of the genes.
The data also showed that some chemicals can influence more than one cellular process. These compounds are more likely to cause side effects and are best avoided. "With our map, we can see these promiscuous compounds earlier and focus on the good ones," noted Piotrowski.
In this work, the researchers focused on studying how drugs affected single mutants, and subsequently, cellular health. In doing so, the investigators could find how specific drugs exerted their effect. This methodology allowed the scientists to look at the influence of one drug, in the context of a whole organism.
Learn more about how researchers use microbes to make new drugs from the following video.
Sources: AAAS/Eurekalert! Via University of Toronto, Nature Chemical Biology