Understanding the Enzymes of the Microbiome
In the avalanche of research being generated on the human microbiome, it’s important to take a step back and think about the many basic facts of the system that we don’t know anything about. One glaring lack of knowledge regards the molecules that associate with the microbial community residing in the gastrointestinal tract – the gut microbiome. Scientists think it’s likely that many of those molecules are enzymes, acting in chemical reactions that we know nothing about, but which are affecting health or disease. A bioinformatics tool has now been developed to get at these mysteries.
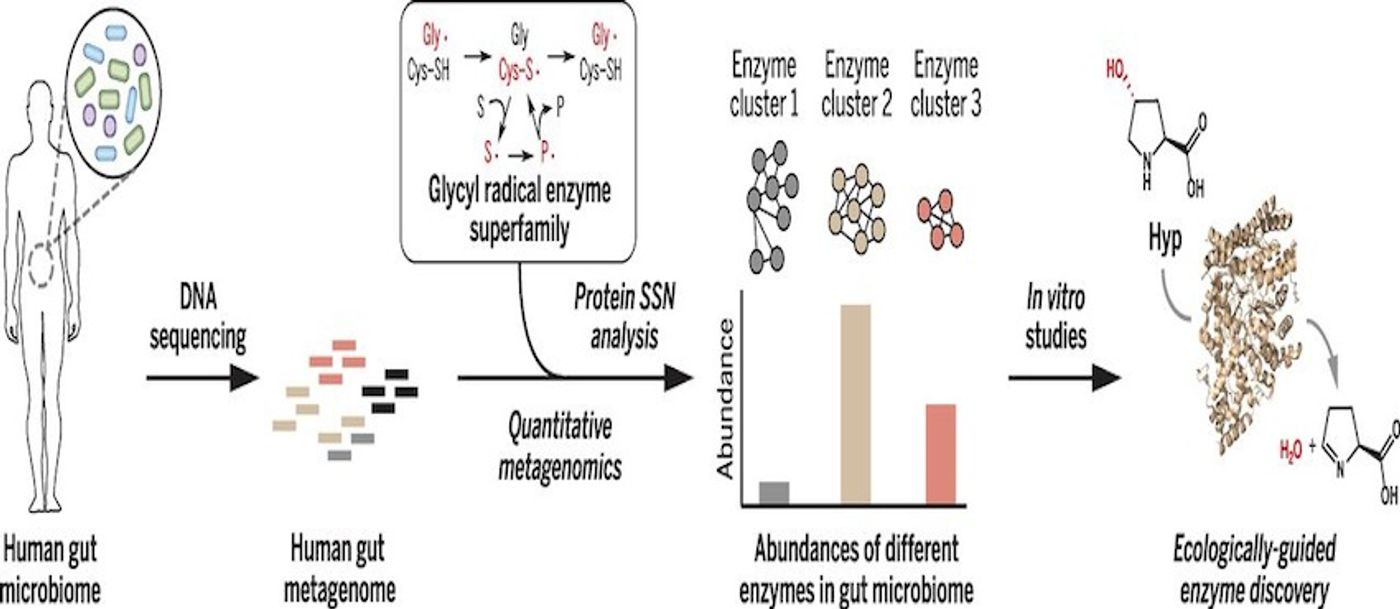
Emily Balskus, the Morris Kahn Associate Professor of Chemistry and Chemical Biology worked with Curtis Huttenhower, an Associate Professor of Computational Biology and Bioinformatics at the Harvard T.H. Chan School of Public Health to create a technique that will aid in the identification and quantification of enzymes present in microbiomes. Their work has been published in Science. In the video above, she gives a talk on the chemicals of the microbial world.
"This uncharacterized enzyme was the second most abundant glycyl radical enzyme in every person sequenced as part of the Human Microbiome Project, and its universal distribution strongly suggested it was doing something important" Balskus said. "It turns out to have a fascinating function. It's allowing microbes to metabolize an amino acid called 4-hydroxyproline, which is a major component of collagen, the most abundant protein in the human body. We've discovered how microbes use this amino acid to grow in the anaerobic environment of the human gut."
Scientists will now be able to progress to new questions by utilizing the information garnered by the new technique, added Balskus.
"Now that we have an appreciation that this activity is highly abundant in this microbial habitat, we can begin to explore a number of new ideas for why it might be present and how it might be affecting the human host and other microbes," she said.
Balskus noted that because of the myriad commonalities between enzymes, it’s incredibly challenging to find novel molecules.
"We were interested in the challenge of distinguishing members of the same enzyme family that have different activities from one another. Members of a human family can be closely related but have very different occupations, and this is also the case for enzyme families.” Balskus explained. “A family of enzymes can be similar in terms of their amino acid sequences, but the individual members of the family have often evolved to perform very different chemical transformations."
Researchers can now utilize this technique to find and characterize novel enzymes, which will help to reveal how the metabolic mechanisms of the microbiome exert an effect on the local environment and beyond.
"I hope this will be an important contribution and step toward tackling this huge problem of uncharacterized proteins in microbial communities," she commented. "This is a problem not just for the human gut microbiome, but for any microbial community. And these communities are literally everywhere on Earth!"
Balskus thinks this new tool could also help uncover how the microbial community living in the human gastrointestinal tract influences health.
"In the future we can start to think about things like comparative analyses," she said. "In this paper we only looked at data from healthy humans, but we'd really like to compare the abundance of enzymes in microbiomes from healthy individuals and patients suffering from various diseases. This could give us a glimpse into how metabolic activities in the gut microbiome might be changing with disease. If there are certain enzymes that are particularly abundant in patients with a given disease, they could be potential therapeutic targets."
Dr. Balskus did a webinar for Labroots in 2015 regarding chemistry of the human microbiota, it’s presented in the following video.
Sources: AAAS/Eurekalert! via Harvard University, Science