With a New Technique, a Mystery About mRNA is Solved
When a gene is active, cellular machinery transcribes that gene into a molecule called messenger RNA (mRNA). A variety of modifications are typically made to mRNA molecules before they are used further by the cell, usually to generate a protein. Scientists have now learned more about a mysterious RNA modification, a methyl group that is found in a certain place. The findings have suggested that this type of methylation is related to antiviral defense mechanisms in cells, and disruptions to the methyl modification may be related to some inflammatory or autoimmune diseases. The work has been reported in Nature.
"We've known since the 1970s that methyl modifications are somehow fundamental to how mRNAs normally work. So it's very gratifying to finally have this insight into its precise role," said senior study author Dr. Samie Jaffrey, a Professor at Weill Cornell Medicine.
The activity of genes can be controlled in many ways, and mRNA molecules can add another layer of regulation, and may promote or inhibit the production of proteins from mRNA molecules. Methyl groups are often involved in this process.
The Jaffrey lab has previously developed ways to identify methyl modifications called methyl adenosine (m6A) tags on mRNA; they can control mRNA stability. There is another common mRNA modification called Cap2, which has been mystery to investigators for many years.
The ends of chromosomes are protected by 'caps' known as telomeres. There are 'caps' of nucleotides that are also found on mRNA molecules, and a methyl group is attached to the first nucleotide base of the mRNA sequence. This initial methyl group is a standard 'Cap1' tip. It protects mRNA from immune processes that eliminate anything that looks like viral mRNA from the cell.
Some mRNA molecules also carry additional methyl groups at the second nucleotide; this is a 'Cap2' methylation. For many years, scientists have been trying to determine why some mRNAs carry Cap2 while others do not. One challenge has been differentiating between Cap1 and Cap2 groups.
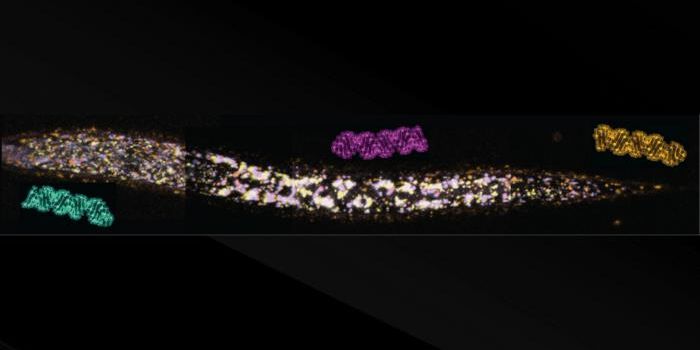
Jaffrey and first study author Vladimir Despic, a postdoctoral researcher, created CLAM-Cap-seq to discern between Cap1 and Cap2. They found that Cap2 can be found on any mRNA, but it only appears after an mRNA molecule has been hanging around the cell for awhile.
Additional work showed that Cap1 helps stop mRNA molecules from stimulating an immune response. Cap2 adds more protection. When cells only carried mRNAs with Cap1, the researchers found that antiviral inflammatory processes started up, even when no virus was present.
Cells have to keep Cap2 levels in check as well. When too much Cap2 was added to mRNA molecules, invading viruses began to infect cells, which could not respond well enough to eliminate the infections.
"We think Cap2 methylation occurs slowly, rather than quickly, in order to reduce the chance it will end up cloaking fast-replicating viral RNAs," Jaffrey said.
Dysfunction in the Cap1/Cap2 process may be involved in some common inflammatory and autoimmune disorders, and the researchers are investigating. When viral infections are difficult to treat, it may also be possible to boost antiviral mechanisms in cells by inhibiting Cap2 .
"We're also studying the possibility of using Cap2 modifications to improve mRNA-based therapeutics, including vaccines, by reducing their inflammatory effects in cells," added Jaffrey.
Sources: Cornell University, Nature