New study finds that the structure of chromosomes may not be as important to transcriptional activity as we once thought
Among scientists, it is widely accepted that the structure of genomes largely controls their transcriptional activity within the cell. Structural features known as chromatin are responsible for this regulation of activity. Chromatin is a tightly wound section of DNA and protein within the nucleus of the cell. How tightly (heterochromatin) or loosely (euchromatin) DNA is wound directly affects the transcriptional activity of that section of DNA. Or does it?
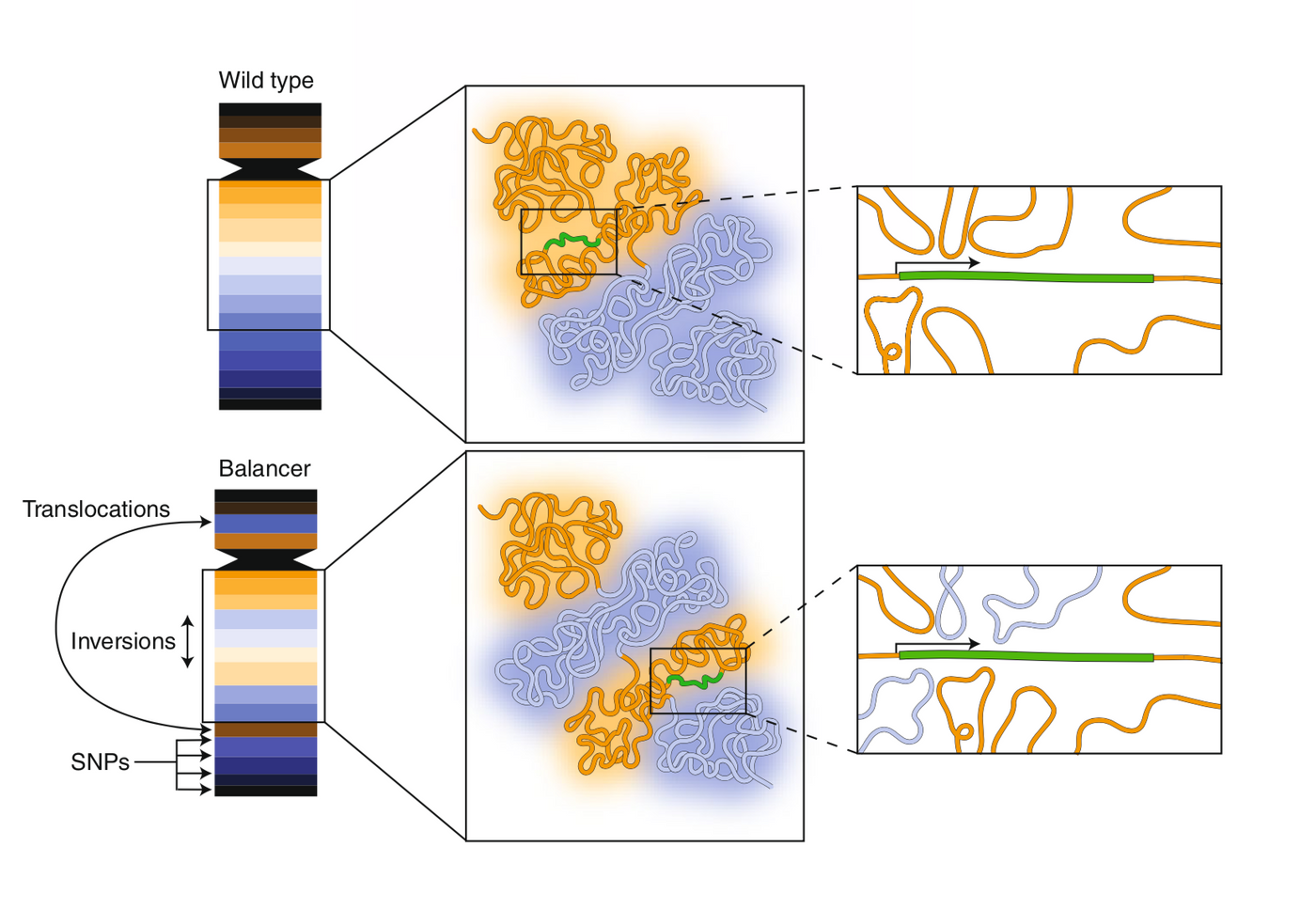
Ghavi-Helm et al. of the Furlong Lab at the European Molecular Biology Laboratory in Heidelburg, Germany recently published findings in Nature Genetics that contradict this widely accepted theory. The team used balancer chromosomes in Drosophila melanogaster (or fruit flies) to look more closely at the relationship between chromatin structures and gene expression. Balancer chromosomes are used to create a population of flies with a mutation in one set of chromosomes that will be maintained throughout several fly generations and will not become lethal to the fly. In this study, the second and third chromosomes were mutated with many inversions, duplications, and deletions. These balancers and the wild-type chromosomes were used to "systematically map allele-specific chromatin topology" and compare it to the corresponding region of the chromosome.
Interestingly, only a subset of genes expression was altered when the chromosome was rearranged, and the topology was effected. According to Ghavi-Helm et al., "these results suggest that many genes are generally impervious to the large-scale topological features of their surroundings." However, some genes were effected in instances where there were far more topological changes, supporting the original theory. Together this study reveals that the structure of the genome is not enough to determine gene transcriptional activity.
Article sources: A genome disconnect, Yad Ghavi-Helm et al.
To learn more about chromatin and its role in gene regulation, watch this video: