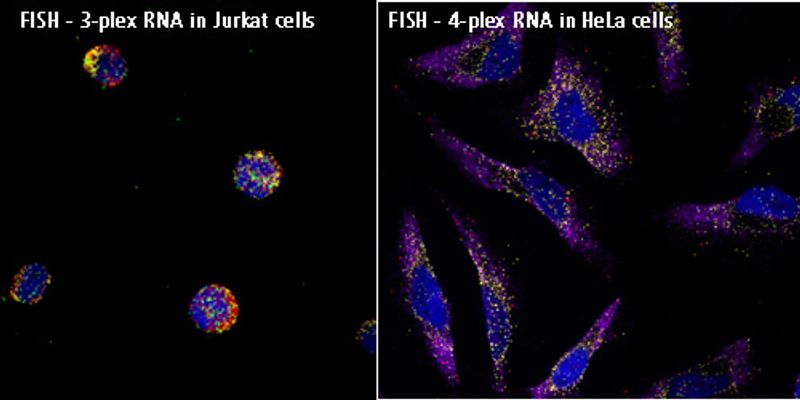
RNA FISH
Fluorescence in situ hybridization - aka FISH is defined as a molecular cytogenetic technique that uses fluorescent probes that bind to only those parts of a nucleic acid sequence with a high degree of sequence complementarity. Developed in the early 1980's, biomedical researchers developed it to localize and detect the absence or presence of specific DNA sequences on chromosomes. Fluorescence microscopy can be utilized to find out where the fluorescent probe is bound to the chromosomes. FISH is often used for finding specific features in DNA for use in genetic counseling, medicine, and species identification. FISH can also be used to detect and localize specific Ribonucleic acid or RNA targets in tissue samples, circulating tumor cells, and cells, assisting in defining the spatial-temporal patterns of gene expression within tissues and cells.
-
Date: November 19, 2020 Time: 12:00am (PDT), 9:00am (CET), 4:00pm (SGT) We present split-FISH, a multiplexed fluorescence in situ hybridization method that leverages a split-probe design to...
Sponsored By: Accelerate Diagnostics